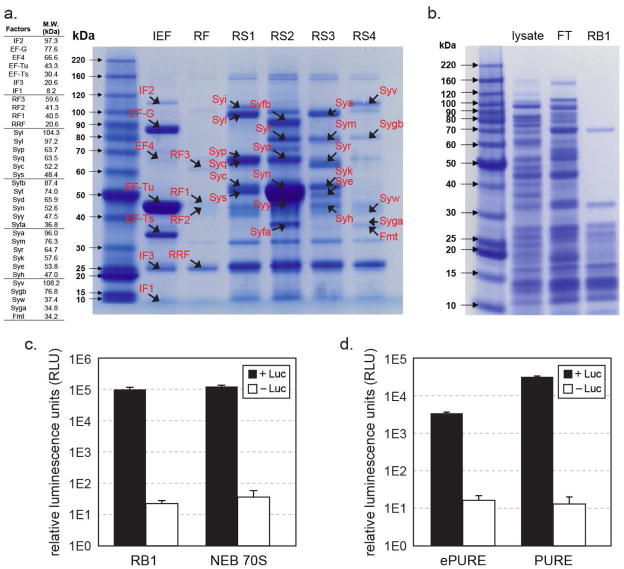
Figure 3. Assessment of purified His-tagged ribosome and translation factors.
(a). A table of translation factors by protein size is listed on the left panel (not including His-tags). The right panel shows eluted fractions of His-tagged IEF, RF, and RS1-4 factors after Ni-NTA purification on Bis-Tris PAGE gel, stained by Coomassie-blue. The expected migration band of each factor is marked on the gel. Though a few of the visible bands may be non-translation factor, Ni-binding proteins (e.g. at 25 kD), presence of translation factors in the elution fraction was validated by mass spectrum (Table 2). Eluted fractions from RF strains contained very faint amount of proteins. Re-examination of the washed fractions showed that RF1, RF2, RRF and RF3 eluted off the Ni-NTA resin consecutively during the wash step. (b). His-tagged ribosome purified from RB1 cell lysate, FT: flow through. (c). Activity comparison of RB1 ribosome that has His6-tags on L7/L12 ribosomal proteins and was purified over Ni-NTA resin with wild-type E. coli 70S ribosome (New England Biolabs) purified by the traditional sucrose gradient method. Ribosomes were compared using an in vitro translation assay producing firefly luciferase, with chemical luminescence plotted on a log scale. Control reactions were conducted under same conditions but without luciferase gene template. Error bars are ± standard deviations, with n=3. (d). Activity of co-purified ePURE factor mix, supplemented with IF1, IF3, ArgRS, GlyRS, demonstrated by in vitro translation of luciferase and luminescence. PURE factor mix prepared by individual factor purifications is also performed under the same conditions as a positive control. Negative control reactions were conducted in reactions without luciferase gene template. Error bars are ± standard deviations, with n=4.