Figure 1. Schematic representation of SNaPshot genotyping.
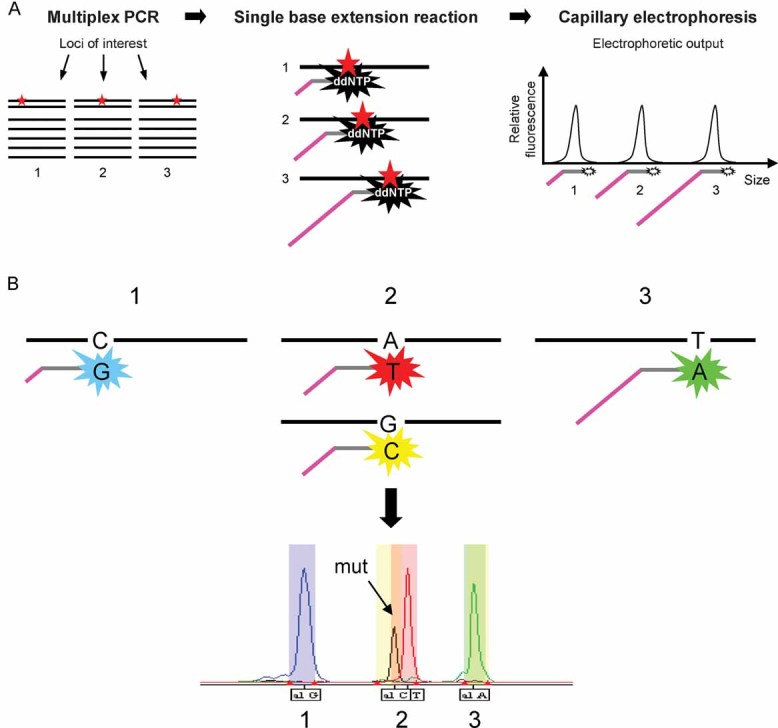
- The SNaPshot system follows a straightforward protocol and uses infrastructure already existent in most clinical laboratories. This method consists of a multiplexed PCR step, followed by a single-base extension sequencing reaction, in which allele-specific probes interrogate loci of interest and are fluorescently labelled using dideoxynucleotides. These probes are designed to have different sizes and are subsequently resolved by electrophoresis and analysed by an automated DNA sequencer. Thus, the identity of each locus is given by the position of its corresponding fluorescent peak in the spectrum, which is dictated by the length of the extension primer.
- Detailed view of the single-base extension reaction. The identity of the nucleotide(s) present at each locus is given by two parameters: the molecular weight and the colour of the fluorescently labelled ddNTPs added to the allele specific probes during the extension step. Thus, mutant and wild-type alleles can be distinguished based on the slightly different positions and on the distinct colours of their corresponding peaks. These two factors are used to establish the bins used for automatic data analysis (described in the Supporting Information).
