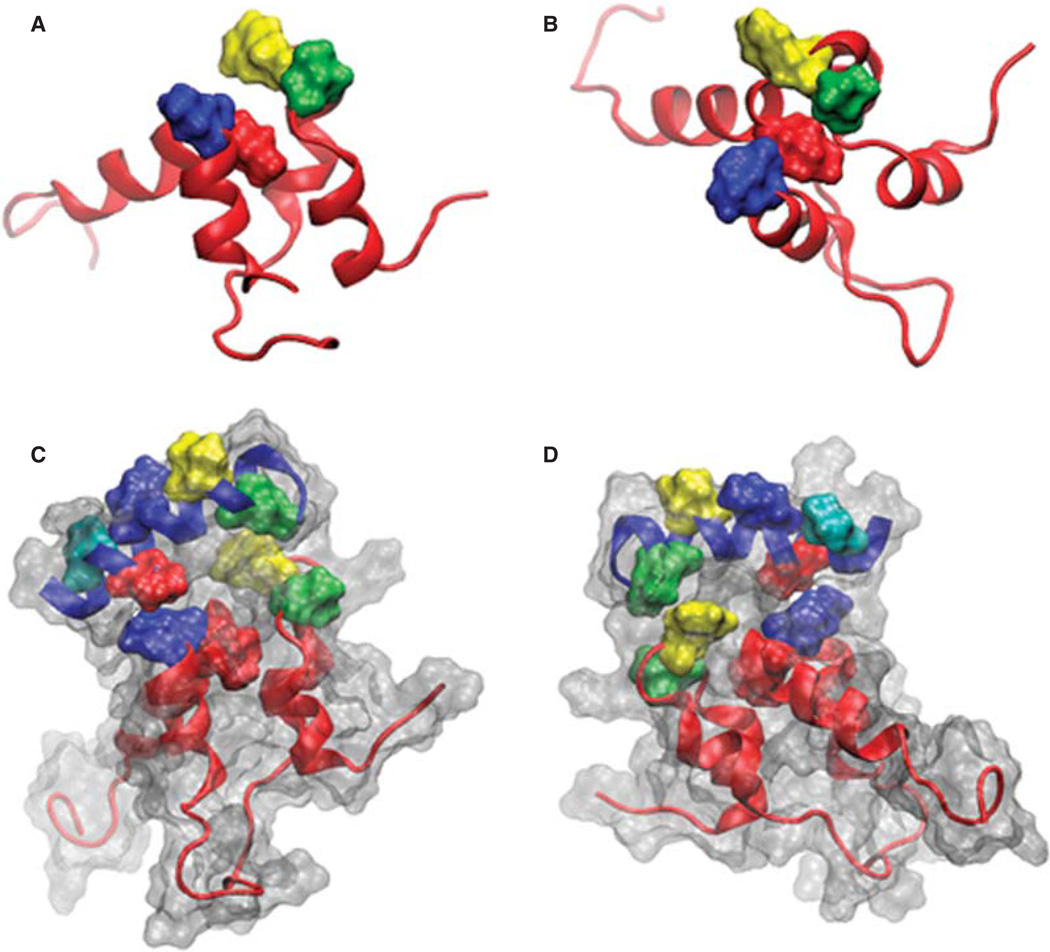
Figure 1.
Images of sAnk-1 alone and the sAnk-157–122:Obsc6322–6339 complex. (A) Top and (B) side views of the hydrophobic hotspot within the binding region of sAnk1, from the 30 ns time frame of the sAnk-1 monomer MD simulations. The backbone is shown in red cartoon representation while the four surface-exposed hydrophobic residues: Val-70 (green), Phe-71 (yellow), Ile-102 (blue), and Ile-103 (red) are shown as surface representations, (C) Front and (D) rear views of the sAnk-157–122:Obsc6322–6339 complex. sAnk-1 is shown in a red cartoon representation along with the four residues shown in panels A and B; obscurin is shown in a blue cartoon representation with L6326 (green), V6328 (yellow), I6332 (blue), V6334 (red) and V6336 (cyan) shown as a surface representation. Full solvent accessible surfaces representations of both proteins are included as white transparent surfaces. Structures for panels C and D were from the 7.7 ns time frame of the 30 ns MD simulation of wild type sAnk1 in complex with Obsc6322–6339, which has an average VDW interaction energy of 1.62 kcal/mol between I102 of sAnk1 and V6334 of obscurin.