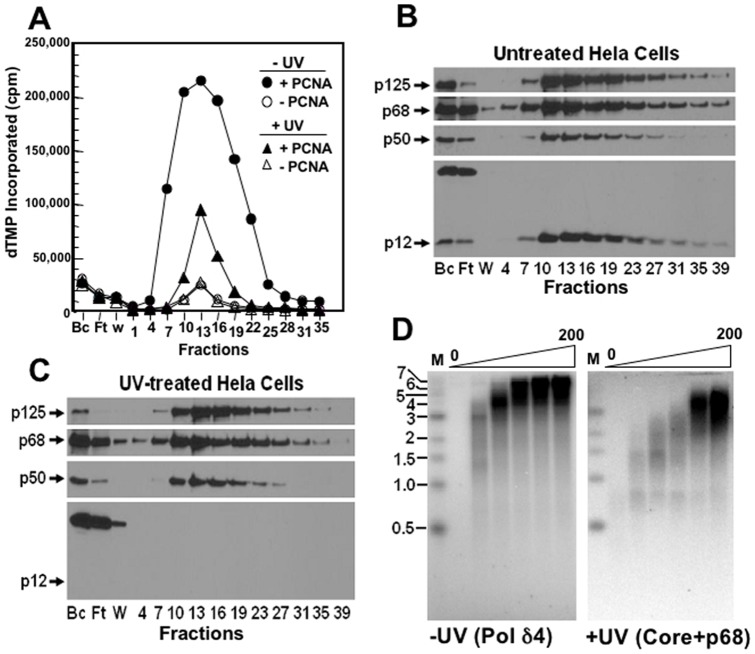
Figure 3. Isolation of Pol δ from UV-treated HeLa cells by immunoaffinity chromatography.
HeLa cells (2×108 cells) were grown and divided into two portions, one of which was treated with UV-C (20 J/m2 at 254 nm) and harvested 4 hour later and the other was used as the control. The UV-treated and control cells were lysed and purified concurrently on two 5-ml columns of anti-p125-agarose. Panel A. Immunoaffinity chromatography: Elution of Pol δ activity. The column fractions were assayed for polymerase activity using poly(dA)/oligo(dT) as the template-primer in the presence (solid circles), and absence (open circles) of PCNA for the untreated cells and for the UV-treated cells (solid triangles, with PCNA; open triangles, without PCNA). The vertical axis indicates the incorporated dTMP in CPMs and the horizontal axis shows the lysate, flow-through, wash, and column fraction numbers. Panel B. The lysate (Bc), flow-through (Ft), wash (W), and column fractions (0.3 ml each) of the immunoaffinity chromatography of the untreated HeLa cells were Western blotted for p12, p125, p50, and p68 (arrows). The lanes from left to right show the column fraction numbers. Panel C. The corresponding fractions from the immunoaffinity chromatography of Pol δ from the UV treated cells were Western blotted for the Pol δ subunits as for Panel B. Panel D. The abilities of the immunoaffinity purified native Pol δ4 (untreated cells) and core+p68 (UV-treated cells) enzymes were tested for their abilities to elongate singly primed M13 DNA (Materials and Methods); images show the gel electrophoresis patterns for product formation. Horizontal bars on the left indicate the positions in kb of DNA markers (“M”). The lanes from left to right show increasing amounts of enzyme (0, 12.5, 25, 50, 100, 200 fmoles).