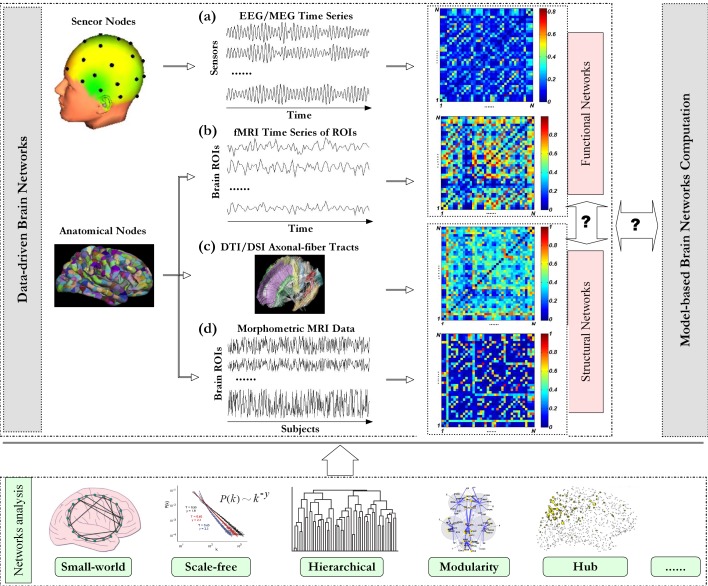
Figure 1.
Schematic paradigm of graph theoretical analysis of brain networks. Investigation on brain networks includes data-driven brain network analysis [5, 6] and model-based brain network computation [42, 43]. With neuroimaging data, different types of data-driven brain networks can be constructed. (a) For EEG/MEG, multichannel time series are measured with an array of sensors, and the scalp region where each sensor located is defined as one node respectively. Pair-wise association between nodes can be quantified with time series at the corresponding locations [44]. For fMRI and DTI/DSI data, regions of interesting (ROIs) are first defined with predefined anatomical templates [6, 14]. (b) The representative fMRI time series of each ROI is extracted from all the voxels in that ROI via a certain way (e.g., by averaging). Pair-wise association between ROIs can be estimated from the representative time series. (c) For DTI/DSI data, pair-wise association can be traced with axonal-fiber tracts [29]. (d) Morphometric variables can be estimated for each ROI from individual structural MRI images, and then multivariate morphometric (e.g., cortical thickness) time series over subjects can be obtained. Pair-wise association between these multivariate time series can be used as a measure of anatomical connectivity [18]. With association matrix constituted by connectivity in all possible node pairs, graph theoretical analysis can be performed to complex brain network. This figure is organized with our data and others [16, 21, 45–47].