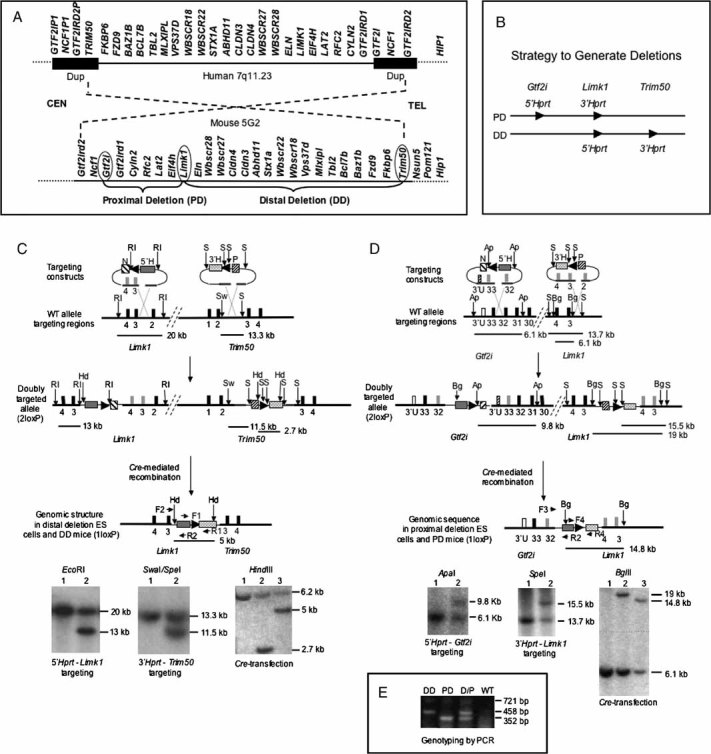
Comparison of WBSCR genomic regions in human and mouse shows conservation of gene order and the area inverted with respect to centromere (CEN) and telomere (TEL).
To generate partial deletion mice that model the WBS deletion, constructs containing loxP sites (arrowheads) and partial Hprt sequences were inserted by homologous recombination at three locations. Cre-mediated recombination between the loxP sites generates the desired deletions.
-
To generate DD mice, a Limk1/neomycin resistance (N)/ 5′-Hprt (5′H) construct was first introduced into HPRT-deficient ES cells. Correctly targeted neomycin-resistant clones were then transfected with the Fkbp6/puromycin resistance (P)/ 3′-Hprt (3′H) construct. Doubly targeted (2loxP) cells were transiently transfected with a Cre-recombinase expression vector. Recombination between the two loxP sites in cis generates a restored Hprt minigene whose expression enabled selection for DD containing ES cells in HAT-medium. Arrowheads indicate the location and orientation of loxP sites; black blocks represent numbered genomic exons; grey blocks symbolize exons in the vector. Restriction sites: EcoRI (RI), SpeI (S), SwaI (Sw), HindIII (Hd). Primer pair F1/R1 amplifies the restored Hprt sequence. Primer pair F2/R2 is used for PCR genotyping of DD. Horizontal lines indicate location and size of restriction fragments.
Lower left: Southern blot confirms correct targeting of 5′-Hprt construct. Lane 1, WT ES cells; lane 2, targeted ES cells. Lower middle: Southern blot confirms correct insertion of 3′-Hprt construct. Lane 1, WT ES cells; lane 2, targeted ES cells. Lower right: Southern blot confirms the Cre-mediated deletion in 1loxP cells. Following HindIII digestion, a PCR amplified fragment within 3′-Hprt was used as probe. Lane 1, WT ES cells; lane 2, 2loxP ES cells; lane 3, 1loxP ES cells. The ∼6.2 kb band in all lanes is due to non-specific cross-hybridization.
To generate the PD mice, the Gtf2i locus was targeted with a 5′-Hprt construct, and subsequent transfection with a Limk1 3′-Hprt construct yielded doubly targeted 2loxP ES cells. All symbols are as in (C) and open blocks represent Gtf2i 3′ UTR sequences (3′U). Restriction sites: ApaI (Ap), BglII (Bg). Primer pair F3/R2 is used to genotype PD mice by PCR. Primer pair F4/R4 amplifies the loxP/Hprt sequence. Lower left: Southern blot confirms correct targeting of 5′-Hprt construct. Lane 1, WT ES cells; Lane 2, targeted ES cells. Lower middle: Southern blot confirms correct insertion of 3′-Hprt construct. Lane 1, WT ES cells; lane 2, targeted ES cells. Lower right: Southern blot confirms Cre-mediated deletion in 1loxP cells. Lane 1, WT ES cells; lane 2, 2loxP ES cells; lane 3, 1lox ES cells.
Multiplex PCR for genotyping of DD, PD and double heterozygous (D/P) mice with primers F2 (for DD), F3 (for PD), and R2 (for both). F4/R4 generate a721 bp fragment from the inserted Hprt sequences, and its absence confirms WT.