Abstract
Here we demonstrate a new regulatory mechanism for tRNA processing in Escherichia coli whereby RNase T and RNase PH, the two primary 3′ → 5′ exonucleases involved in the final step of 3′-end maturation, compete with poly(A) polymerase I (PAP I) for tRNA precursors in wild-type cells. In the absence of both RNase T and RNase PH, there is a >30-fold increase of PAP I-dependent poly(A) tails that are ≤10 nt in length coupled with a 2.3- to 4.2-fold decrease in the level of aminoacylated tRNAs and a >2-fold decrease in growth rate. Only 7 out of 86 tRNAs are not regulated by this mechanism and are also not substrates for RNase T, RNase PH or PAP I. Surprisingly, neither PNPase nor RNase II has any effect on tRNA poly(A) tail length. Our data suggest that the polyadenylation of tRNAs by PAP I likely proceeds in a distributive fashion unlike what is observed with mRNAs.
INTRODUCTION
In Escherichia coli, diverse processing pathways employing RNase E, RNase P, polynucleotide phosphorylase (PNPase) and RNase II are used to generate pre-tRNA species from primary transcripts (1–5). Subsequently, the pre-tRNAs undergo additional maturation at either both their 3′- and 5′-ends or at their 3′-ends only (3–10) to generate functional tRNAs. It has been shown that RNase T, RNase PH, RNase D, RNase BN are the four 3′ → 5′ exoribonucleases that primarily participate in 3′-end maturation of tRNAs and many other stable RNAs (6,7,9), although RNase BN has endonucleolytic activity in vivo and is also called RNase Z (11–13).
Interestingly, E. coli can use some incompletely processed stable RNAs (rRNAs) without significant effects on their growth phenotype (8,14–16), but tRNAs are an exception. All tRNAs, which account for the major fraction of stable RNAs in the cell (17), must be completely processed at their 3′-termini before they can be aminoacylated and used in protein synthesis (18). Although the 3′-ends of some tRNA precursors are probably matured by a preferred set of exonucleases (3,5,9), it is thought that the majority of the 3′-end tRNA maturation is carried out by RNase T and RNase PH with only minor contributions by RNase D and RNase BN (9).
Taken together these results present a comprehensive overview of the processing of primary tRNA transcripts into functional species. However, the observation of polyadenylated tRNAs in the absence of both RNase T and RNase PH (16,19) was rather unexpected, particularly since polyadenylation in E. coli by poly(A) polymerase I (PAP I) has been almost exclusively characterized in relation to mRNAs (20–23). Furthermore, sequencing analysis of hisR, cysT and leuX transcripts has shown that a significant fraction (∼20–33%) of the transcripts have short poly(A) tails in a RNase PH single mutant (4,5). Interestingly, the fraction of normal pre-tRNAs (not defective based on nucleotide sequence) with poly(A) tails was considerably higher than previously observed for other transcripts (mRNAs and rRNA) in E. coli (24,25).
Although no poly(A) tails have been detected on mature tRNAs or 5S rRNA in wild-type E. coli, Li et al. (26) proposed a model in which the primary function of polyadenylation was to identify and present defective tRNA processing intermediates for recycling through degradation pathway(s) that are part of a general quality control process. The evidence for poly(A)-dependent degradation of a mutant tRNATrp (26) and various mRNAs (22,24,27) provided support for this hypothesis, but it did not explain the presence of polyadenylated pre-tRNA transcripts in the rph-1 mutant that were not defective (4,5).
Similarly, it has been suggested that the shorter poly(A) tails observed on many stable RNA precursors resulted from degradation of longer poly(A) tails by exoribonucleases such as polynucleotide phosphorylase (PNPase), RNase II or RNase R (19). However, inactivating either PNPase or RNase II did not change the length of poly(A) tails associated with leuX transcripts (5). Taken together, these data suggested a potentially more significant role for the observed polyadenylation of pre-tRNAs in E. coli.
Here, we present a detailed analysis of 50 out of 86 tRNA primary transcripts along with 5S rRNA that demonstrates a second function for RNase T and RNase PH beyond their role in tRNA 3′-end maturation. Specifically, both enzymes reduce or block the addition of short poly(A) tails by PAP I on tRNAs, with RNase T being more effective than RNase PH. Furthermore, polyadenylation induces rapid degradation of 5S rRNA precursors but only a small fraction of tRNA precursors. Rather the majority of polyadenylated precursors are slowly converted into mature species, most likely by RNase D and/or RNase BN. The data are consistent with the 2- to 4-fold drop in the pool of aminoacylated tRNA species along with a concomitant >2-fold increase in cell generation time in an Δrnt rph-1 double mutant. In contrast, charged tRNA levels and growth rate improved significantly in a ΔpcnB Δrnt rph-1 triple mutant. Furthermore, a small number of tRNAs (7/86) are resistant to polyadenylation even in the absence of both RNase T and RNase PH. Of particular interest is the fact that PAP I apparently acts on tRNAs substrates in a distributive manner compared to a more processive mechanism for mRNAs.
MATERIALS AND METHODS
Bacterial strains and plasmids
The E. coli strains used in this study were all derived from MG1693 (rph-1 thyA715) (E. coli Genetic Stock Center, Yale University). This strain contains no RNase PH activity and has reduced expression of pyrE because of a single nucleotide frameshift in the rph gene (28). A rph+ derivative was constructed by transducing a P1 lysate grown on E. coli C600 into MG1693 and selecting for faster growing isolates on minimal medium. Several independent transductants were sequenced to confirm the presence of the wild-type rph coding sequence. One such isolate was designated SK10153 (thyA715). SK9124 [rph-1 thyA715/pBMK11(pcnB+/CmR)] and SK10148 (Δrnt::kan rph-1 thyA715) have been previously described (2,24). A P1 lysate grown on SK10148 was used to transduce SK10153 (thyA715) and SK10019 (pnpΔ683 rph-1 thyA715) to construct SK10592 (Δrnt::kan thyA715) and SK10609 (Δrnt pnpΔ683 rph-1 thyA715), respectively. SK10575 (Δrnb Δrnt rph-1 thyA715) was made by transduction with P1 phage grown on CMA201 (Δrnb::tetR rph-1 thyA715) (29) into SK10148 (Δrnt::kan rph-1 thyA715).
The ΔpcnB::aac(3)-IV (apramycin, AprR) deletion/substitution allele in SK4465 was obtained using the method of Hamilton et al. (30). Briefly, the pcnB coding sequence starting from amino acid six after the UUG translation start codon until two amino acids upstream of the translation stop codon was replaced by the aac(3)-IV apramycin resistance cassette obtained from plasmid pSET152 (Genbank Accession No. 414670). SK10593 [Δrnt ΔpcnB::aac(3)-IV] and SK10020 [Δrnt ΔpcnB::aac(3)-IV rph-1 thyA715] were constructed by transducing SK10592 and SK10148, respectively, with P1 grown on SK4465. Plasmid pBMK58(rnt+/ApR) was constructed by cloning the rnt coding sequence containing its own promoter into pWSK29 (31) at the BamHI–PstI sites. A PCR fragment containing the rnt coding sequence was amplified using primer pairs RNT-BAMH (175 nt upstream of ATG start codon) and RNT-PST (74 nt downstream of TAA) using Phusion™ high fidelity DNA polymerase (NEB). SK10589 (Δrnt rph-1/pBMK58) was constructed by transforming pBMK58 into SK10148.
Northern analysis, dot blots and poly(A) sizing assay
The details of the poly(A) sizing assay, RNA isolation, Northern analysis and dot blots have been described previously (32). All strains were grown to Klett 50 (∼108 cells/ml, No. 42 green filter) for isolation of steady-state RNA. For RNA half-life experiments rifampicin (500 µg/ml) and nalidixic acid (40 µg/ml) were added to cultures at Klett 50 prior to harvesting the cells at selected times. For PAP I induction, IPTG (350 µmol) was added to cultures at Klett 50 and cells were collected at the specified time points. All RNAs were quantified by measuring the OD260 using a Nanodrop (2000C) apparatus (ThermoFisher) and were initially analyzed on an 1% agarose gels in TAE (Tris–acetic acid-EDTA) to check their integrity. RNA concentrations were normalized to a wild-type control (32). For Northern analysis, total RNA (12 µg for tRNAs, 2.5 µg for 5S rRNA and 5 µg for lpp mRNA per lane) was separated in 6% polyacrylamide gels containing 8 M urea in TBE (Tris–Borate-EDTA buffer) and transferred to a positively charged nylon membrane (Nytran® SPC, Whatman®). Poly(A) sizing assays were carried out using 20% polyacrylamide gels.
Western blotting
Cells from 50 ml of exponentially growing cultures were harvested by centrifugation at 5000 rpm for 5 min at 4°C in a Beckman–Coulter™ (Avanti™ J-25) centrifuge. Cell pellets were resuspended in 300 µl of CellLytic™ B Bacterial Cell Lysis/Extraction Reagent (Sigma®) containing protease inhibitors (SIGMAFAST™ Protease Inhibitor Cocktail, EDTA free, Sigma® and 1 mM PMSF) and stored on ice. Cells were disrupted by sonication (Misonix, S-4000) and crude extracts were clarified by centrifugation in a microcentrifuge at full speed for 15 min at 4°C. Protein samples were quantified by Bradford assay using a Nanodrop (2000C) apparatus. Protein samples (125 µg) were separated on SDS-polyacrylamide (10%) gel and electrotransferred to PVDF membrane (Immobilon™-P, Millipore) using a Bio-Rad Mini-Protean 3 gel apparatus. Antibodies against PAP I (kindly provided by Grzegorz Wegrzyn, University of Gdañsk, Poland) were initially incubated in 750 µg of cell lysate obtained from a pcnB deletion strain for 1 h and used to probe the membrane using Pierce® ECL Western Blotting substrate. The protein bands were detected and quantified using a G:Box (Syngene).
Determination of aminoacylation levels of tRNAs
All the strains were grown with shaking to Klett 50 at 37°C. Cells from 5 ml of culture were collected at 4°C and total RNA was isolated under acidic condition essentially as described by Varshney et al. (33). In one set of RNA samples (3 µg each), tRNA was deacylated by treatment with 0.5 M Tris–HCl (pH 9.0) in a total volume of 40 µl for 30 min at 37°C. The RNA was precipitated at −20°C by adding 1/10th volume of 3 M sodium acetate (pH 5.2), 1 µl of GlycoBlue™ (Ambion) and 2.5 volume of 100% ethanol. The RNA was pelleted by centrifugation for 15 min at 4°C. The pellets were dissolved in the original volume of 10 mM sodium acetate (pH 4.5) and 1 mM Na2EDTA. Both untreated and Tris treated RNA samples (3 µg each) were mixed with an equal volume of loading buffer and separated on a 6.5% acid urea gel in the cold room as described by Varshney et al. (33) with following modifications. The electrophoresis was carried out at 250 V for ∼8 h using a gel (19 cm long) until the xylene cyanol dye front was ∼3–4 cm from the bottom of the gel. The RNA was transferred to a positively charged nylon membrane as described previously (32).
RNase H treatment
Total RNA (5–12 µg) was treated with RNase H (USB) in presence of oligo-d(T)20 to remove poly(A) tails as described previously (34).
Probe selection, hybridization of membranes and quantification of data
Oligonucleotide probes (Supplementary Table S1) were selected to hybridize to the mature sequences of all tRNAs and 5S rRNA. Various tRNA isotypes were differentiated based on differences in their mature sequences. Oligonucleotide probes were 5′-end labeled using T4 polynucleotide kinase (NEB) and γ-32P-ATP (Perkin Elmer) as described elsewhere (32). In most cases, the same membrane was probed multiple times after stripping the previous probe as previously described (32). The lpp mRNA was detected by probing with 32P-labeled lpp DNA fragments as described previously (23). All Northern blots and poly(A) sizing gels were scanned using a PhosphorImager (GE Healthcare, Storm™ 840). All data were quantified using ImageQuant TL 5.2 software (GE Healthcare).
RT-PCR cloning and sequencing of 5′–3′ ligated transcripts
The 5′- and 3′-ends of tRNAs and 5S rRNA were identified by cloning and sequencing the reverse transcription-PCR products obtained from 5′- to 3′-end-ligated circular RNAs following the methods described previously (32). The 5′–3′ junctions of the cDNAs were amplified with a pair of gene-specific primers using GoTaq®Green Master Mix (Promega).
RESULTS
The level of short poly(A) tails (<10 nt) increases dramatically in an RNase T RNase PH double mutant
Since some of the tRNA precursors in an RNase T RNase PH double mutant as well as 5S and 23S rRNA precursors in RNase T single mutants have been shown to contain short poly(A) tails (16,19,35), we hypothesized that PAP I was significantly involved in stable RNA metabolism. Accordingly, we carried out dot blot experiments to determine the total poly(A) level in a RNase T RNase PH double mutant. We were surprised to find an ∼50% reduction in total poly(A) level compared to a wild-type control (data not shown). However, since dot blots do not detect poly(A) tails <15 nt in length, we performed a poly(A) sizing assay, which can visualize poly(A) tails as short as a single nucleotide.
The polyadenylation profiles of a wild-type control and a RNase PH single mutant were almost identical, although a small but reproducible increase in total poly(A) level spanning all size ranges was observed in an rph-1 strain (Figure 1, lanes 2 and 3; Table 1). In contrast, in an RNase T single mutant there was ∼2.5-fold decrease in the amount of longer poly(A) tails (>10 nt) accompanied by a 4-fold increase in the level of short poly(A) tails (≤10 nt) (Figure 1, lane 4 and Table 1) compared to both the wild-type and rph-1 strains with the majority of the increase occurring in the range of 1–6 nt (Figure 1, lane 4). In the absence of both RNase T and RNase PH, there was a dramatic increase in the level of 4- to 9-nt long poly(A) tails compared to the Δrnt single mutant (Figure 1, lane 5). Overall, there was ∼30-fold increase in the amount of short poly(A) tails (≤10 nt) in the RNase T RNase PH double mutant compared to the wild-type control (Figure 1, lanes 2 and 5; Table 1). Interestingly, the level of longer poly(A) tails (>10 nt) remained almost unchanged in the RNase T RNase PH double mutant compared to the RNase T single mutant (Figure 1, lanes 4 and 5; Table 1). Overexpression of RNase T through the introduction of pBMK58 (a 6- to 8-copy number plasmid carrying the rnt under the control of its own promoter) into the RNase T RNase PH double mutant led to a polyadenylation profile that was identical to the wild-type control (data not shown).
Figure 1.
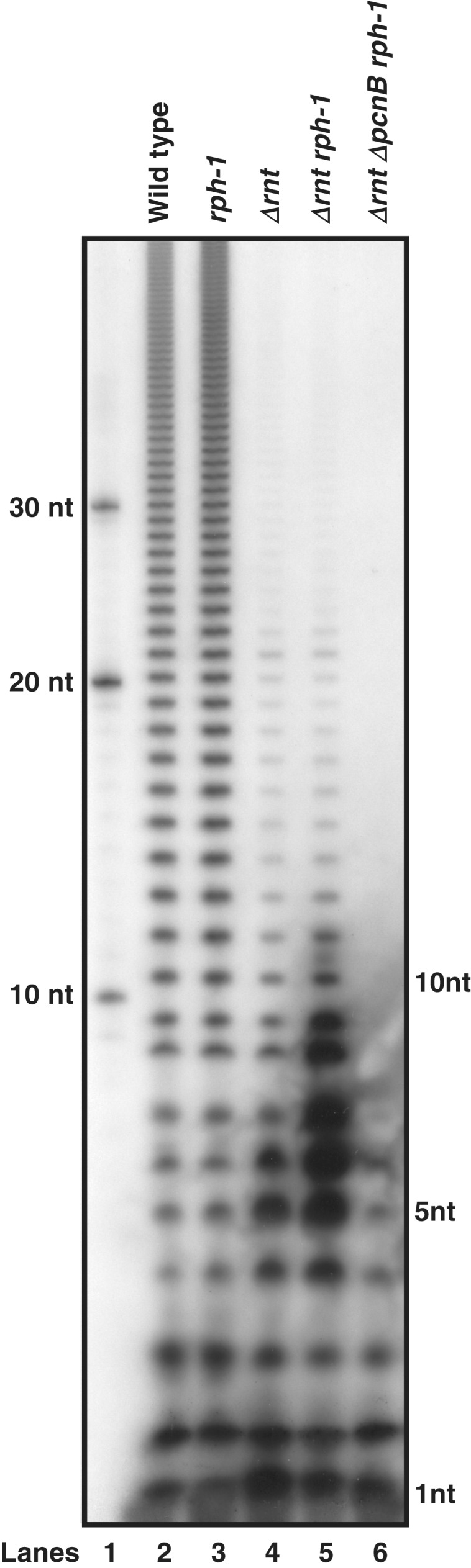
Distribution of poly(A) tails in various E. coli strains. Total RNA (15 µg/lane) from exponentially growing cultures were 3′-end-labeled with [32P]-pCp in presence of T4 RNA ligase followed by digestion with RNase A and RNase T1. The digested samples were separated on a 20% PAGE as described previously (32). It should be noted that RNase A cleaves after C and U residues and RNase T1 cleaves after G residues. As a result, 3′ poly(A) tails are protected and are increased in length by 1 nt by the addition of 32pCp. Thus, the in vivo size of corresponding band is noted to the right. Lane 1, 5′-32P-labeled d(A) size standards; lane 2, SK10153; lane 3, MG1693; lane 4, SK10592; lane 5, SK10148; lane 6, SK10020. The genotype of each strain is listed at the top of each lane.
Table 1.
Poly(A) tail distribution in various strains
| Strain | Genotype | Relative amounts of poly(A) tailsa |
|
|---|---|---|---|
| <10 nt | >10 nt | ||
| SK10153 | thyA715 | 1 | 1 |
| MG1693 | rph-1 thyA715 | 1.3 ± 0.2 | 1.4 ± 0.3 |
| SK10592 | Δrnt thyA715 | 4 ± 1 | 0.4 ± 0.1 |
| SK10148 | Δrnt rph-1 thyA715 | 30 ± 5 | 0.5 ± 0.1 |
| SK10020 | ΔpcnB Δrnt rph-1 thyA715 | 0.6 ± 0.2 | ND |
aThe level of poly(A) tails was determined by separately integrating all bands >10 nt and ≤10 nt as shown in Figure 1 using ImageQuant TL 5.2 software (see ‘Materials and Methods' section). The pixel counts for the respective group of bands in the wild-type control were set at one.
ND = not detected.
The short poly(A) tails observed in RNase T mutants are synthesized by PAP I
PAP I (encoded by pcnB) is the major polyadenylating enzyme in E. coli (22,24,25), although PNPase also adds heteropolymeric tails to a limited extent (36,37). As shown in Figure 1 (lane 6), deletion of pcnB led to the disappearance of all the longer (>10 nt) poly(A) tails as well as the vast majority of the smaller ones (≤10 nt), indicating that the tails were added primarily by PAP I in both the RNase T single (data not shown) and the RNase T RNase PH double mutant. The few remaining small tails (≤6 nt) in the Δrnt ΔpcnB rph-1 triple mutant (Figure 1, lane 6) most likely arose from either the biosynthetic activity of PNPase or chromosomally encoded As that were present at the 3′-ends of RNA breakdown products (36).
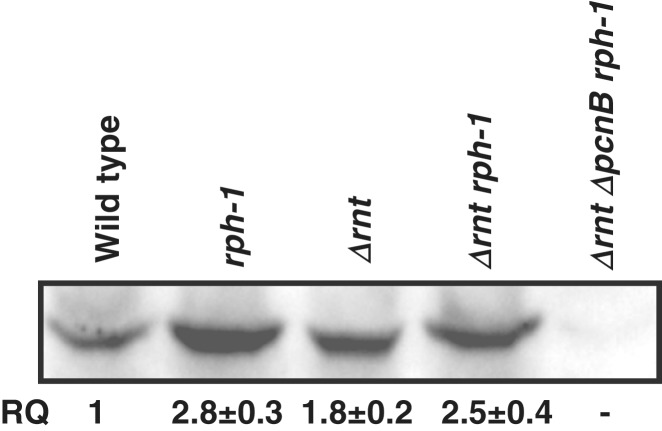
In order to determine if the 30-fold increase in the level of short poly(A) tails in the RNase T RNase PH double mutant was due to increased levels of PAP I protein, western-blot analysis was carried out. PAP I protein levels increased between 1.8- and 2.8-fold in all the mutants compared to the wild-type control (Figure 2). The increase in PAP I protein level in the rph-1 single mutant was consistent with the increase in the level of poly(A) tails across all size ranges compared to the wild-type control (Figure 1, lanes 2 and 3; Table 1). However, the increases in PAP I protein in the Δrnt single and Δrnt rph-1 double mutants were less than what was seen in the rph-1 single mutant, even though there were 4- and 30-fold increases in short poly(A) tails in the respective strains (Figure 1, lanes 4 and 5; Table 1).
Figure 2.
Western blot analysis of PAP I levels in SK10153 (wild-type), MG1693 (rph-1), SK10592 (Δrnt), SK10148 (Δrnt rph-1) and SK10020 (ΔpcnB Δrnt rph-1) strains. RQ (relative quantity) represents the average of three independent determinations. The PAP I level in SK10153 was set at 1.
Increased levels of short poly(A) tails result from polyadenylation of tRNA and 5S rRNA precursors
The tRNA concentration in exponentially growing E. coli has been estimated to be ∼0.5 mM (2–3 × 105 molecules per cell), which constitutes up to 20% of the total RNA (38–40). Although previous observations have shown that some tRNAs and rRNAs precursors contained short poly(A) tails in the absence of either RNase PH or RNase T or both (4,5,8,9,16), these results were never quantitatively analyzed to determine if the polyadenylation profiles of specific species changed in various mutants as shown in Figure 1. We hypothesized that the extensive increase in poly(A) levels in Δrnt rph-1 double mutant resulted from extensive polyadenylation of unprocessed 3′-ends of tRNAs and 5S rRNA, which were normally substrates for RNase T and PH (9,19).
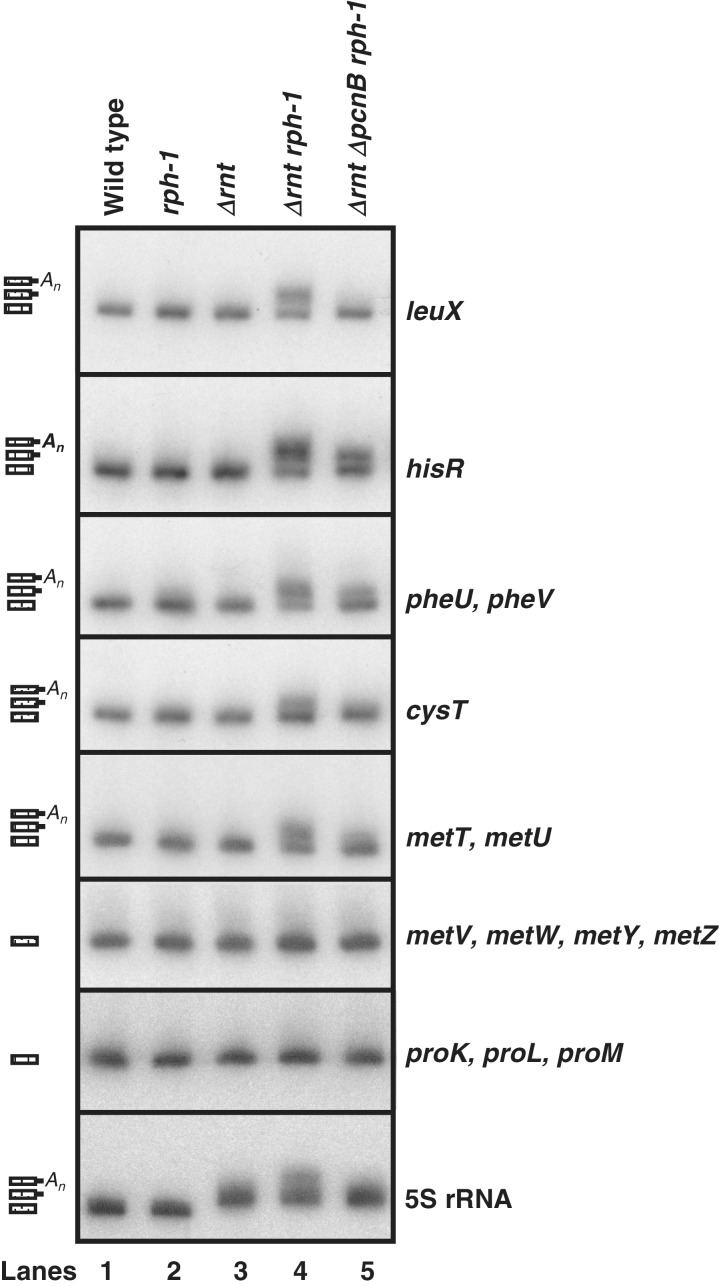
Accordingly, we quantitatively analyzed a series of tRNAs, as well as 5S rRNA, in strains defective in various combinations of RNase T, RNase PH and PAP I using Northern blots. In an initial study of 14 tRNA species, the absence of either RNase PH or RNase T alone had no significant effect on maturation (Figure 3, lanes 1–3). In contrast, in the absence of both RNase T and RNase PH, there were significant amounts of higher molecular weight species presumed to be the immature precursors in the case of leuX, hisR, pheU, pheV, cysT, metT and metU (Figure 3, lane 4). Surprisingly, in the case of the three proline tRNAs (proK, proL and proM) and the metf1 and f2 isotypes (metV, metW, metY and metZ) inactivation of RNase T and RNase PH did not result in the accumulation of any detectable immature species (Figure 3, lane 4). In the case of 5S rRNA, the absence of RNase T alone led to a quantitative shift to a larger species (Figure 3, lane 3), in agreement with previous work that has shown that RNase T was essential for removing the three extra nucleotides from the 3′-terminus of the p5S rRNA species (16).
Figure 3.
Northern analysis of tRNAs and 5S rRNA in various E. coli strains. Total RNA was separated on 6% PAGE/8 M urea, transferred to a positively charged nylon membrane and probed with transcript specific probes as described in the ‘Materials and Methods’ section. Lane 1, SK10153; lane 2, MG1693; lane 3, SK10592; lane 4, SK10148; lane 5, SK10020. The genotypes of the strains used are noted above each lane. The deduced structures of the processing intermediates (mature, immature and polyadenylated, from the bottom of each panel) of each transcript are shown to the left.
The immature species observed with the leuX, hisR, pheU, pheV, cysT, metT and metU tRNAs and 5S rRNA in the Δrnt rph-1 double mutant were not discrete in size and appeared to vary in length by a small number of nucleotides (Figure 3, lane 4). Since previous work indicated that some immature tRNA and 5S rRNA precursors were polyadenylated (19), we also analyzed the pattern of precursors in a Δrnt rph-1ΔpcnB triple mutant. In some cases, inactivation of PAP I activity led to an almost complete loss of the immature species (leuX and cysT, Figure 3, lane 5). With other tRNAs, (hisR, pheU, pheV, metT and metU) some immature species were still observed but they were shorter than those seen in the Δrnt rph-1 double mutant (Figure 3, lane 5). In the case of 5S rRNA, the polyadenylated species in the Δrnt rph-1 mutant (Figure 3, lane 4) disappeared completely, but the intermediate containing three extra nucleotide at the 3′-terminus remained (Figure 3, lane 5).
In order to provide further support for our hypothesis that the >30-fold increase in poly(A) levels observed in the Δrnt rph-1 double mutant resulted primarily from polyadenylation of tRNA precursors, we expanded our study to include 50 of the 86 E. coli tRNA genes (Table 2). Overall 24 distinct tRNA isotypes as well as one rRNA encoded by 58 different genes were quantitatively examined (Table 2). Only seven tRNA transcripts out of 58 genes studied (12%) were not affected by the loss of both RNase T and RNase PH (Table 2). In contrast, the precursors for the vast majority of tRNAs (43 of 50) and 5S rRNA accumulated in the Δrnt rph-1 double mutant. The level of tRNA precursor accumulation varied from as little as 20% to as high as 71% compared to the wild-type control (Table 2). Furthermore, the levels of precursor tRNAs were significantly reduced (8–59%) in the ΔpcnB Δrnt rph-1 triple mutant compared to the Δrnt rph-1 double mutant supporting a direct role for PAP I.
Table 2.
Effect of absence of RNase T and RNase PH on selected tRNAs and rRNA
| tRNA isotype/rRNA | No. of Loci | Gene(s) | Percentage of immature transcriptsa |
Presence of poly(A) tailsb | |
|---|---|---|---|---|---|
| Δrnt rph-1 | Δrnt ΔpcnB rph-1 | ||||
| Arg2 | 4 | argQ, argV, argY, argZ | 45 ± 3 | 14 ± 2 | Yes |
| Asn | 4 | asnT, asnU, asnV, asnW | 27 ± 4 | 8 ± 2 | Yes |
| Cys | 1 | cysT | 44 ± 0 | 31 ± 2 | Yes |
| Glu2 | 4 | gltT, gltU, gltV, gltW | 26 ± 3 | 17 ± 0 | Yes |
| His | 1 | hisR | 71 ± 2 | 59 ± 1 | Yes |
| Leu1 | 4 | leuT, leuQ, leuP, leuV | 41 ± 1 | 20 ± 3 | Yes |
| Leu2 | 1 | leuU | 20 ± 2 | 12 ± 2 | Yes |
| Leu3 | 1 | leuW | 54 | 44 | Yes |
| Leu4 | 1 | leuZ | 38 | 29 | Yes |
| Leu5 | 1 | leuX | 61 ± 1 | 30 ± 5 | Yes |
| Lys | 6 | lysQ, lysT, lysV, lysW, lysY, lysZ | 24 | 9 | Yes |
| Met f1, Met f2 | 4 | metZ, metW, metV, metY | 0 | 0 | No |
| Met m | 2 | metT, metU | 58 ± 0 | 41 ± 3 | Yes |
| Phe | 2 | pheU, pheV | 67 ± 3 | 45 ± 5 | Yes |
| Pro1, Pro2, Pro3 | 3 | proK, proL, prom | 0 | 0 | No |
| Ser1 | 1 | serT | 50 | 23 | Yes |
| Ser3 | 1 | serV | 56 | 42 | Yes |
| Ser5 | 2 | serW, serX | 36 | 9 | Yes |
| Val1 | 5 | valU, valX, valY, valT, valZ | 53 | 48 | Yes |
| Val2A, Val2B | 2 | valV, valW | 53 | 38 | Yes |
| 5S | 8 | rrfA, B,C,D,E,F,G,H | 100 ± 0 | 100 ± 0 | Yes |
aRepresents the fraction of immature species out of total (mature, immature, and polyadenylated species, see Figure 3) RNA species detected. Data, with standard deviations, represent the average of at least three independent experiments.
bPoly(A) tails were scored as being present if the immature species observed in the Δrnt rph-1 strain were shortened in the ΔpcnB Δrnt rph-1 triple mutant.
Interestingly, the absence of PAP I had a variable effect on the reduction of the fraction of immature tRNA species (both polyadenylated and non-polyadenylated). Although the fraction of immature species for most of the tRNAs studied was reduced between 1.1- and 2.7-fold, for some pre-tRNA species (Arg2, Asn and Ser5) the fraction of immature species was reduced between 3.2- and 4-fold compared to the Δrnt rph-1 double mutant. In the case of 5S rRNA, the level of precursors was 100%, as expected based on previously published work (16). The seven tRNAs that did not use either RNase T or RNase PH for their 3′-end maturation were also not substrates for PAP I (Table 2).
Polyadenylated tRNA and 5S rRNA precursors have significantly altered half-lives
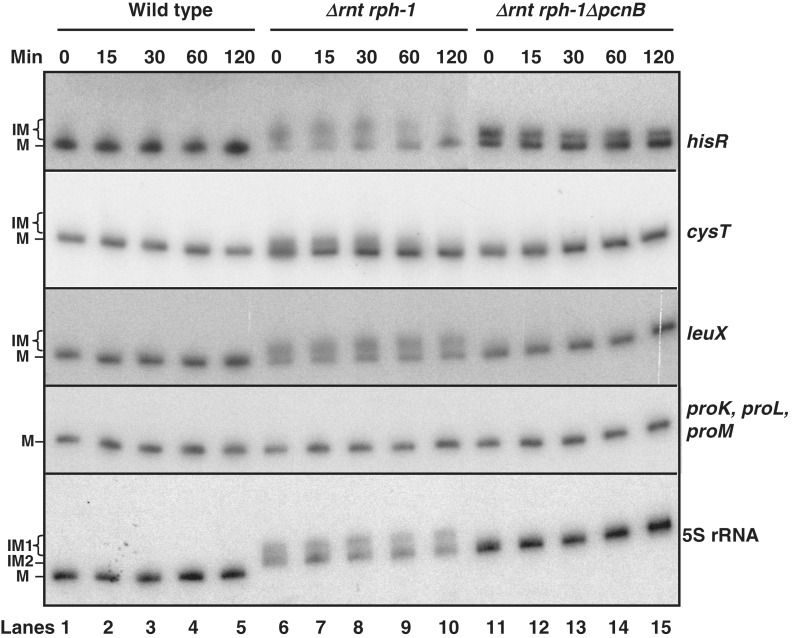
Since it has been argued that polyadenylation functions as part of the cell's RNA quality control pathway (26), we reasoned that the polyadenylated species observed in Figure 3 should have significantly shorter half-lives than their comparable mature species. This idea would be consistent with previous reports showing that polyadenylated mRNAs and small regulatory RNAs (sRNAs) have shorter half-lives compared to the non-polyadenylated species (22,24,27,41–45).
Accordingly, we measured the half-lives of 16 tRNAs and 5S rRNA (Table 3). The mature species for all the tRNAs tested had half-lives >120 min in the wild-type control. In fact, the level of most of the mature tRNAs actually increased steadily during each experiment (Figure 4, lanes 1–5; Table 3 and Supplementary Figure S1). Similar increases in mature tRNA levels after rifampicin addition have been previously observed (38). All the tRNAs tested in the Δrnt rph-1 double mutant had a distinct band corresponding to immature and/or polyadenylated precursor species (IM), with the exception of the three proline tRNAs (proK, proL and proM) where only the mature species (M) was detected (Figure 4). The amount of the immature and polyadenylated tRNA precursors decreased steadily and had measurable half-lives, while the level of mature species increased steadily (Figure 4, lanes 6–10; Table 3 and Figure S1). These results suggested that although some of the immature and/or polyadenylated species might be degraded, a significant fraction were slowly converted into mature species.
Table 3.
Half-lives of tRNAs and 5S rRNA in various genetic backgrounds
| Transcripts | Half-life (min) |
||
|---|---|---|---|
| Wild-type | Δrnt rph-1 | ΔpcnB Δrnt rph-1 | |
| cysT | |||
| IM | NP | 90 ± 10 | NP |
| Ma | ND (↑) | ND (↑) | ND (↑) |
| hisR | |||
| IM | NP | 80 ± 5 | 100 ± 5 |
| M | ND (↑) | ND (↑) | ND (↑) |
| leuT, leuQ, leuP, leuV | |||
| IM | NP | 77 ± 10 | NP |
| M | ND (↑) | ND (↑) | ND (↑) |
| leuW | |||
| IM | NP | 153 | NC |
| M | ND (↑) | ND (↑) | ND (↑) |
| leuZ | |||
| IM | NP | 170 | NP |
| M | ND (↑) | ND (↑) | ND (↑) |
| leuX | |||
| IM | NP | 133 ± 10 | NP |
| M | ND (↑) | ND (↑) | ND (↑) |
| gltT, gltU, gltV, gltW | |||
| IM | NP | 70 | NC |
| M | ND (↑) | ND (↑) | ND (↑) |
| proK, proL, proM | |||
| IM | NP | NP | NP |
| M | ND | ND | ND |
| 5S rRNAb | |||
| IM-1 | NP | 100 ± 8 | NP |
| IM-2 | NP | 120 ± 10 | ND (↑) |
| M | ND (↑) | NP | NP |
aThese tRNA transcripts are mostly polyadenylated in an Δrnt rph-1 strain. bIM-1 is polyadenylated, IM-2 is not polyadenylated in 5S rRNA.
Upward arrow (↑) indicated the band intensities increased up to 120 min.
IM = immature transcripts, M = mature transcripts, NP = not present, NC = no change, ND = not determined.
Figure 4.
Northern blots to determine the half-lives of various tRNA and 5S rRNA transcripts. Total RNA from SK10153 (wild-type), SK10148 (Δrnt rph-1) and SK10020 (Δrnt ΔpcnB rph-1) was isolated at times (minutes after rifampicin addition) indicated at the top of the blot and was separated on 6% PAGE, transferred to a positively charged nylon membrane and probed with transcripts specific probes as described in the ‘Materials and Methods’ section. The blot was scanned using a STORM 840 PhosphoImager (GE Healthcare) and the intensities of the bands were quantified using ImageQuant TL software. The values (log of the percentage of transcript remaining) were plotted as a function of time (Supplementary Figure S1) to calculate each half-life (Table 3). M, IM, IM1 and IM2 are as described in the legend to Table 3.
Surprisingly, the conversion of the immature species appeared to be faster in the absence of PAP I, since the non-polyadenylated tRNA precursors for majority of the tRNAs in the ΔpcnB Δrnt rph-1 mutant were not observed beyond 0-min time point (70 s after rifampicin addition) with the exception of hisR (Figure 4, lanes 11–15). Overall, the level of mature species in the ΔpcnB Δrnt rph-1 triple mutant steadily increased during the experiments, similar to what was observed for the wild-type control (Figure 4, lanes 11–15; Table 4).
Table 4.
Percentage of tRNA aminoacylation in various strains
| tRNA isotypes/genes | Percentage of aminoacylated tRNAs in |
||||
|---|---|---|---|---|---|
| Wild-type | rph-1 | Δrnt | Δrnt rph-1 | ΔpcnB Δrnt rph-1 | |
| Arg2/argQ, argV, argY, argZ | 71 ± 5 | 73 ± 2 | 71 ± 5 | 33 ± 1 | 58 ± 4 |
| His/hisR | 76 ± 1 | 70 ± 5 | 70 ± 5 | 18 ± 4 | 36 ± 4 |
| Leu5/leuX | 91 ± 1 | 89 ± 1 | 88 ± 1 | 38 ± 2 | 79 ± 3 |
| Metm/metT, metU | 85 ± 1 | 82 ± 2 | 80 ± 2 | 33 ± 3 | 56 ± 5 |
| Cys/cysT | 60 ± 3 | 52 ± 3 | 54 ± 3 | 26 ± 2 | 37 ± 2 |
| Lys/lysQ, lysT, lysV, lysW, lysY, lysZ | 70 ± 3 | 71 ± 3 | 69 ± 3 | 30 ± 2 | 54 ± 3 |
| Pro/proK, proL, proM | 75 ± 1 | 78 ± 1 | 74 ± 1 | 74 ± 2 | 75 ± 2 |
The percentage of aminoacylated tRNAs were determined as described in Figure 5. The numbers presented are the average of three independent determinations.
The level of mature 5S rRNA species increased slightly in the wild-type strain during half-life experiment (Figure 4, lanes 1–5; Table 3 and Supplementary Figure S1). In contrast, the levels of both the immature species in the Δrnt rph-1 mutant decreased indicating their instability (Figure 4, lanes 6–10, Supplementary Figure S1). The polyadenylated species IM1 decreased at a faster rate with a half-life of ∼100 min compared to the non-polyadenylated species IM2 with a half-life of ∼120 min (Table 4). However, the level of the immature species in Δrnt ΔpcnB rph-1 mutant increased under similar experimental conditions (Figure 4, lanes 11–15, Supplementary Figure S1).
The absence of polyadenylation improves aminoacylation levels and growth rate in an Δrnt ΔpcnB rph-1 triple mutant
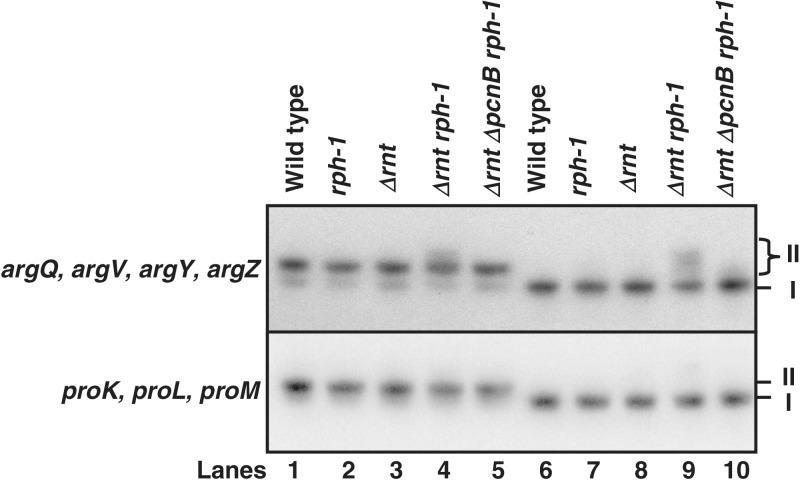
The extent of aminoacylation of mature tRNAs is an important factor in maintaining the normal growth rate in any organism. Considering that a significant fraction of most tRNAs were immature and polyadenylated in the Δrnt rph-1 double mutant (Table 2), we assumed that these species were not charged, resulting in poor growth. In contrast, since the inactivation of PAP I led to decreases in immature tRNA species along with concomitant increases in mature tRNAs (Table 2 and Figure 4), we predicted an increased level of aminoacylation and a shorter generation time in the triple mutant compared to the Δrnt rph-1 double mutant. To test this hypothesis, we measured in vivo aminoacylation of selected tRNAs using the method of Varshney et al. (33). This method allows the isolation of tRNAs from exponentially growing cells and the separation of charged and uncharged tRNA species on polyacrylamide acid urea gels (Figure 5).
Figure 5.
Representative Northern blots showing the determination of the percentage of aminoacylated tRNAArg2 and tRNAPro. Total RNA isolated from SK10153 (wild-type), MG1693 (rph-1), SK10592 (Δrnt), SK10148 (Δrnt rph-1) and SK10020 (Δrnt ΔpcnB rph-1) were untreated (lanes 1–5) or treated with 0.5 M Tris (pH 9) to chemically deacylate tRNAs (lanes 6–10) and were separated using acid urea polyacrylamide gel as described in Materials and Methods. The position marked with I indicated uncharged form of tRNA. The position marked with II indicated either charged and/or high molecular weight immature tRNAs [with or without poly(A) tails]. The net extent of aminoacylation (lanes 1–5) for band II was calculated as percentage of total counts (bands I + II) minus the percentage of high molecular weight species of total counts (bands I + II) after Tris treatment in lanes 6–10 (33). The aminoacylation levels of all tRNAs studied are reported in Table 4.
The levels of charged tRNAs in the wild-type strain were found to be in the range between 60% and 91% (Table 4 and Figure 5). These results were consistent with previous reports on aminoacylation levels of several tRNAs using same method (33,46–49). As predicted, all six tRNA isotypes transcribed from 15 distinct genes that were affected by the loss of RNase T and RNase PH (Table 2), showed 2- to 4-fold reductions in aminoacylation levels in the Δrnt rph-1 double mutant compared to the wild-type control (Table 4). In contrast, the percentage of charged tRNA increased almost 2-fold for most tRNAs in the ΔpcnB Δrnt rph-1 triple mutant compared to the Δrnt rph-1 double mutant (Table 4). Since the three proline tRNAs (proK, proL, proM) were not affected by the loss of either RNase T and/or RNase PH, their charging levels were comparable in all strains tested (Table 4 and Figure 5).
With regard to generation times, inactivation of RNase PH had no significant effect (Table 5), while the loss of RNase T activity led to small but significant increase in generation time (Table 5). In contrast, in the absence of both RNase T and RNase PH the generation time of the double mutant increased over 2-fold (25 versus 56 min, Table 5). The generation times of the ΔpcnB and ΔpcnB rph-1 mutants were identical and were only marginally slower compared to the wild-type or rph-1 single mutant strains (Table 5). However, although the Δrnt single mutant and the Δrnt ΔpcnB double mutant had somewhat similar growth rates, the generation time of the ΔpcnB Δrnt rph-1 triple mutant improved significantly compared to the Δrnt rph-1 double mutant (Table 5). Overexpression of either RNase PH or RNase T from multicopy plasmids [pPP1 (rph+ ApR) and pBMK58 (rnt+ ApR), respectively, restored the generation time of the Δrnt rph-1 double mutant to the wild-type levels (data not shown).
Table 5.
Generation times of various strains in Luria broth
| Strains | Genotype | Generation time (min)a |
|---|---|---|
| SK10153 | thyA715 (wild-type) | 25 ± 1 |
| MG1693 | rph-1 thyA715 | 27 ± 1 |
| SK10592 | Δrnt thyA715 | 38 ± 2 |
| SK10148 | Δrnt rph-1 thyA715 | 56 ± 2 |
| SK10591 | ΔpcnB thyA715 | 30 ± 1 |
| SK4465 | ΔpcnB rph-1 thyA715 | 30 ± 1 |
| SK10593 | ΔpcnB Δrnt thyA715 | 35 ± 2 |
| SK10020 | ΔpcnB Δrnt rph-1 thyA715 | 48 ± 2 |
aGeneration times were measured at 37°C and represent the average of at least two independent experiments.
RNase T prevents the polyadenylation of mature tRNA transcripts
Having implicated RNase T and RNase PH in the regulation of tRNA and 5S rRNA polyadenylation levels, we next determined the polyadenylation profile and exact location of poly(A) tail additions for selected single copy tRNAs such as hisR (Figure 6), cysT (Figure 6) and leuX (data not shown). In the case of the hisR and cysT pre-tRNAs, there are one to two additional nucleotides downstream of their mature 3′-termini (Figure 6), which arise from RNase E endonucleolytic cleavages of polycistronic transcripts (1,2,4). In contrast, the monocistronic leuX transcript is processed at the 3′-terminus primarily by PNPase leaving between 1- and 3-nt downstream of the CCA terminus (5).
Figure 6.
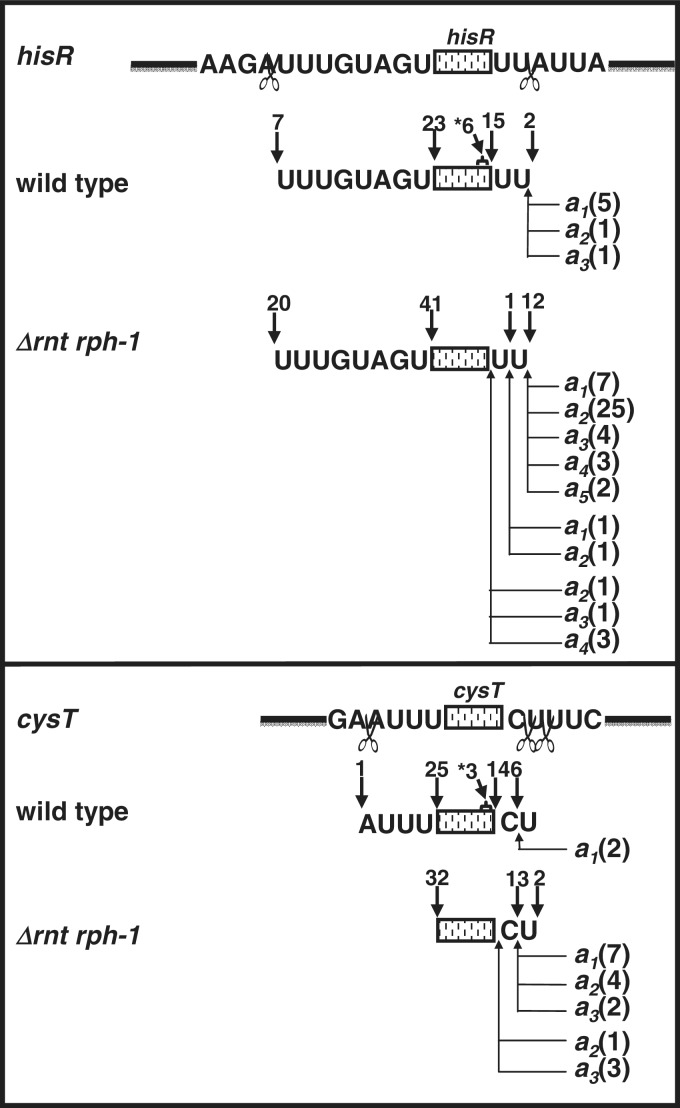
Identification of 5′- and 3′-ends of hisR and cysT transcripts in the wild-type strain (SK10153) and the Δrnt rph-1 double mutant (SK10148) by sequencing of cDNA clones obtained from 5′- to 3′-self-ligated transcripts. Schematic presentations of hisR and cysT tRNAs (rectangles) with the 5′ upstream and 3′ trailer sequences are shown directly above respective tRNA clones. The endonucleolytic cleavages (open scissors) upstream of 5′ and downstream of 3′ mature end (CCA terminus) are as previously identified (1,2,4). The 5′- and 3′-ends (without non-templated a's) are shown as inverted arrows above the sequences. The 3′-ends with non-templated a's are shown as bent upward arrows below the sequences. The source of some a residues in hisR (italicized a, see text) could not be unequivocally determined. The numbers directly above each arrow represent the number of independent clones identified with a respective 5′- or 3′-end. The numbers in parenthesis represent the number of independent clones identified with non-templated a residues. Some of the tRNA transcripts are truncated at 3′-ends by 5–16 nt from CCA terminus (indicated by a star).
We have previously shown that ∼66% of the hisR, cysT and leuX transcripts cloned and sequenced from an rph-1 strain using 5′- to 3′-end ligated transcripts had immature 3′-ends (4,5). About 20–33% of these transcripts also contained poly(A) tails up to 5 nt in length, but none of them were located at the mature 3′-terminus. Using this same method, the number of hisR, cysT and leuX transcripts with immature 3′-ends in the wild-type control (SK10153) was only ∼30%, but between 8% and 23% had poly(A) tails (Figure 6; data not shown). Surprisingly, ∼20% (6/30) of the hisR, ∼12% (3/26) of cysT (Figure 6) and ∼17% (3/18) of leuX (data not shown) transcripts were found to have 3′ truncated sequences (5–16 nt upstream of the CCA terminus) without having poly(A) tails.
In the Δrnt rph-1 double mutant, all of the sequenced hisR, cysT and leuX transcripts (119 total) had immature 3′-ends (Figure 6; data not shown). Furthermore, the percentage of transcripts with poly(A) tails of up to 5 nt in length increased to 80% (48/61) for hisR and 53% (17/32) for cysT (Figure 6). For the majority of the transcripts, the poly(A) tails were at least 1–2 nt downstream of the mature CCA termini (Figure 6). The origin of the single adenine 2-nt downstream of the CCA determinant (a1 in Figure 6) in the hisR clones could not be unequivocally determined because of the presence of a chromosomally encoded A at that location (Figure 6). Interestingly, some of the sequenced transcripts (5/61 for hisR and 4/32 for cysT) had poly(A) tails added immediately following the CCA determinant, which had not been previously seen in either wild-type (Figure 6) or rph-1 strains (3,4). Again, none of the poly(A) tails added to the CCA determinant were >5 nt. It should also be noted that although >50% of the clones were mature tRNAs in the wild-type control, no mature tRNAs were isolated from the Δrnt rph-1 double mutant (Figure 6).
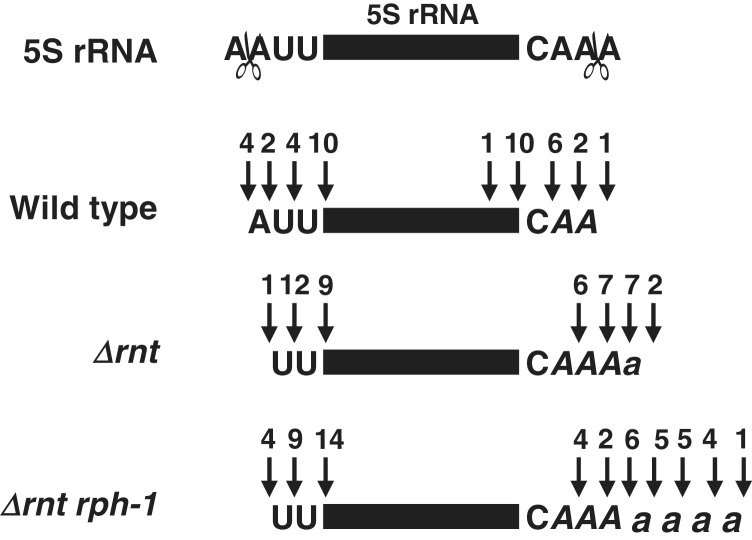
We also sequenced 5S rRNA species derived from wild-type, Δrnt and Δrnt rph-1 strains (Figure 7). This stable RNA is generated endonucleolytically by RNase E cleavages of a larger 9S rRNA precursor (50), creating a p5S RNA species that contains an extra 3 nt at each end (51) (Figure 7). Although the 5′ maturation process is still unknown, the 3 nt at the 3′-end are removed by RNase T (16). Fifty percent (10/20 clones) of the transcripts in the wild-type strain had both 5′ and 3′ mature ends (Figure 7). The rest of the clones had an additional 1–3 nt at both ends. As expected, all the transcripts derived from the Δrnt single mutant (22/22 clones) had immature 3′-ends, although 41% (9/22 clones) had mature 5′-ends (Figure 7). Additionally, all of the transcripts had poly(A) tails that were 1–4 nt in length. While the first three As following C (A) may have been encoded, at least 9% (2/22 clones, small a) of the transcripts had poly(A) tails that were post-transcriptionally added. The number of post-transcriptionally added tails increased to 56% (15/27 clones, small a) and the length of the poly(A) tails increased from 4 to 7 nt in the Δrnt rph-1 double mutant (Figure 7). It should also be noted that none of the pre-tRNAs (with or without poly(A) tails) and pre-5S rRNAs were defective based on their primary nucleotide sequences.
Figure 7.
Identification of 5′- and 3′-ends of 5S rRNA transcripts in the wild-type (SK10153), Δrnt (SK10592) and the Δrnt rph-1 double mutant (SK10148) strains by sequencing of cDNA clones obtained from 5′- to 3′-self-ligated transcripts. Schematic presentation of 5S rRNA (rectangle) with the 5′ upstream and 3′ trailer sequences are shown at the top. The endonucleolytic cleavages (open scissors) upstream of 5′ and downstream of 3′ mature end are as previously identified (49). The numbers directly above each arrow represent the number of independent clones identified with a respective 5′- or 3′-ends. The letters (A, a) indicate nucleotides that were potentially (A) or actually (a) added by poly(A) polymerase.
Poly(A) tails at the 3′-ends of tRNAs are inherently short
Previously we have identified poly(A) tails >17 nt associated with various mRNAs using an oligo(dT)17 primed RT-PCR method (24,25,36,37). Unfortunately, this method does not allow one to determine the actual length of poly(A) tails and, more importantly, it discriminates against detection of poly(A) tails <15 nt. In contrast, RT-PCR cloning using 5′- to 3′-end ligated transcripts is theoretically capable of detecting poly(A) tails of any length, but we were concerned that transcripts with poly(A) tails >5 nt might not ligate efficiently, resulting in the detection of only smaller poly(A) tails (Figures 6 and 7). Furthermore, it was also possible the short poly(A) tails were the result of very low in vivo PAP I levels (37).
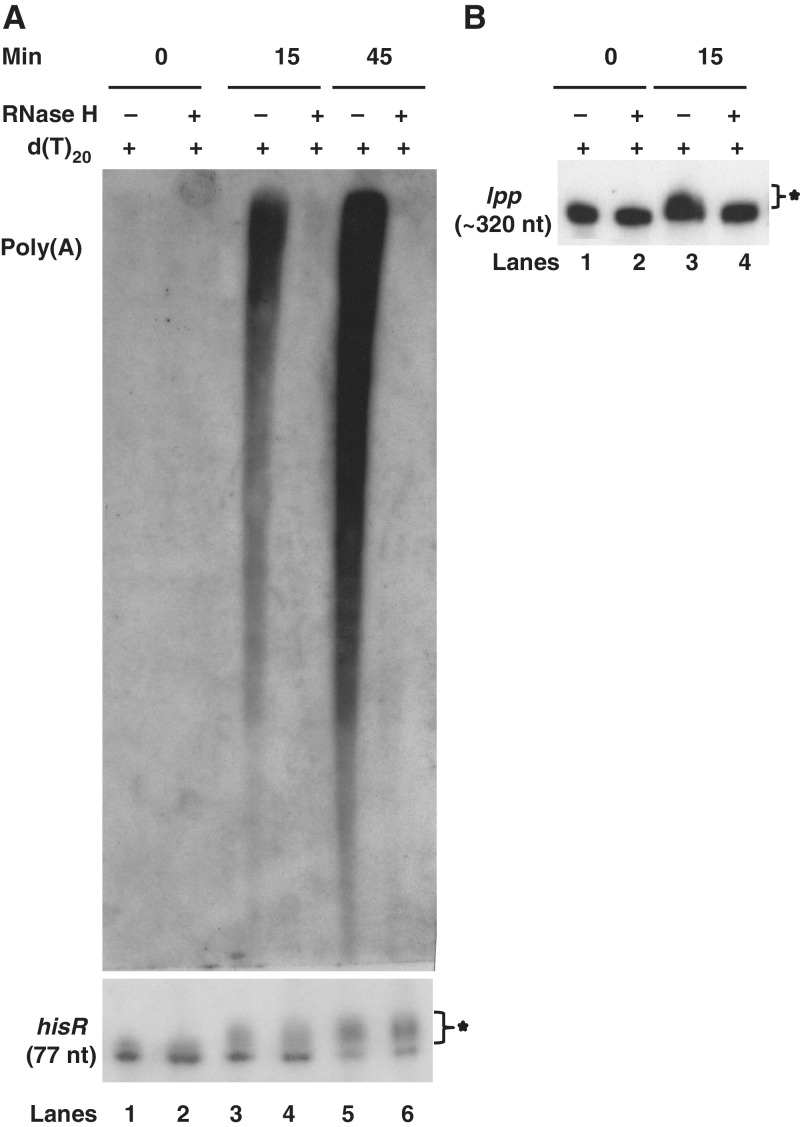
Accordingly, in order to determine if the poly(A) tails added to tRNAs were inherently short, we used an alternative approach in which poly(A) tails >10–15 nt were removed by treating total RNA with RNase H in the presence of oligo(dT)20 (34). This method has been reliably employed by many laboratories to estimate the length of poly(A) tails in both prokaryotes and eukaryotes (34,52,53). In order to overcome the limitation of inherently low PAP I levels (37), we induced PAP I expression from a controlled expression plasmid [pBMK11/CmR, (24)] with IPTG for up to 45 min, circumstances under which we had previously detected long poly(A) tails on the lpp mRNA (24). As shown in Figure 8A, the total poly(A) level increased dramatically during the course of PAP I induction (lanes 3 and 5). Furthermore, pretreatment of the RNA with RNase H in presence of oligo(dT)20 led to an almost complete loss in detectable poly(A) tails (Figure 8A, lanes 4 and 6).
Figure 8.
Characterization of poly(A) tails associated with tRNAs (A) and mRNAs (B) using northern analysis. Total RNA isolated from SK9124 (rph-1/pBMK11) after 0, 15 and 45 min of IPTG (350 µM) induction was either treated (+) or not treated (−) with RNase H in presence of oligo(dT)20 and separated on 6% PAGE as described in the Materials and Methods. The blots were probed with either oligod(dT)20 (to detect poly(A) tails) or probed for hisR, or lpp. A star indicates polyadenylated transcripts. The length of mature species is noted below each transcript.
Subsequently, the blot was reprobed with hisR and cysT specific probes. After 15 min of PAP I induction, ∼25–35% of both the transcripts showed an increase in size (∼5–10 nt) (Figure 8A, lane 3 and data not shown) indicating the addition of short poly(A) tails. Furthermore, the fraction of larger tRNA transcripts increased >50% at the longer time of induction (Figure 8A, lane 5, data not shown). However, the larger transcripts were unaffected by the pretreatment with RNase H (hisR, Figure 8A, lanes 4 and 6; data not shown) indicating that the poly(A) tails on these transcripts were too short to hybridize to oligo(dT)20. As a control we examined the poly(A) tails associated with the lpp mRNA, since it has been shown that they increase in length after 15 min of PAP I induction (24). In fact, we observed a diffuse band of lpp transcripts following 15 min of PAP I induction (Figure 8B, lane 3). As expected, the band became more discrete after RNase H treatment such that it was similar to the bands from the uninduced strains (Figure 8B, lanes 1, 2 and 4).
PNPase and RNase II do not regulate tRNA poly(A) tail length
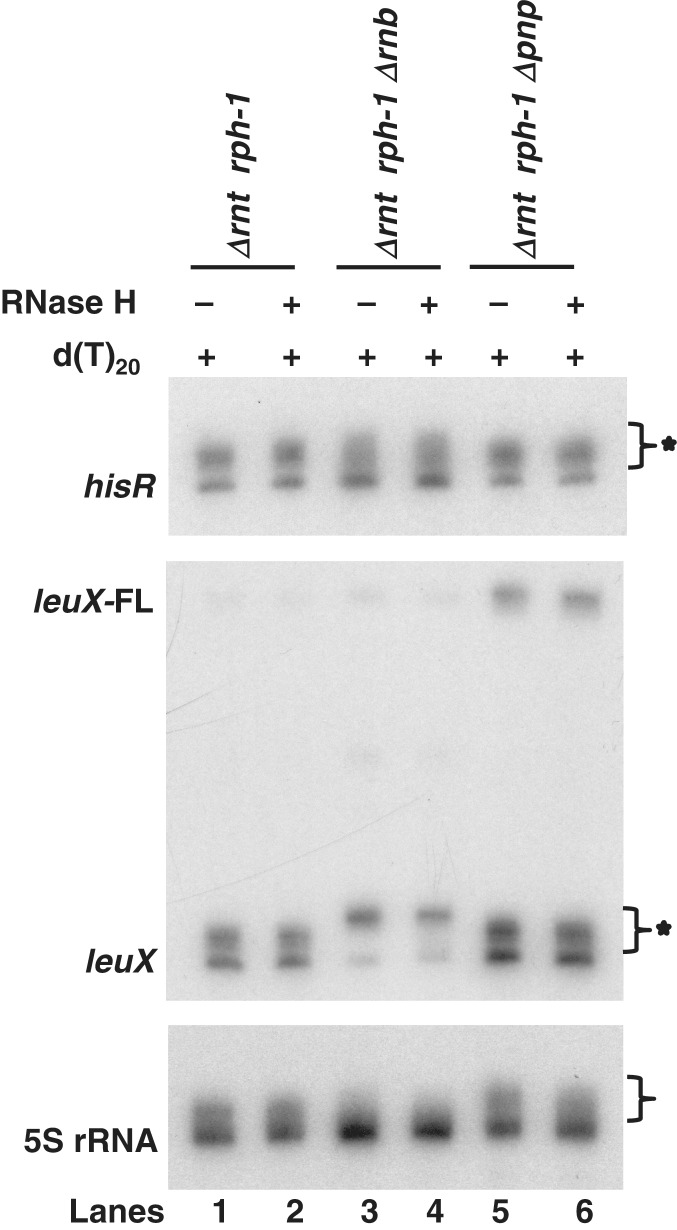
Li et al. (19) suggested that the short poly(A) tails they observed on tRNAs in the absence of RNase T or RNase PH arose from the shortening of longer tails by 3′ → 5′ exonucleases such as PNPase or RNase II. To test this hypothesis, we carried out a poly(A) sizing assay on Δrnt rph-1, Δrnt Δrnb rph-1 and Δrnt Δpnp rph-1 strains where we expected a shift to longer poly(A) tails compared to what was observed in the Δrnt rph-1 double mutant (Figure 1, lane 5). In fact, there was a significant increase in the level of both short and long poly(A) tails in Δrnt Δrnb rph-1 and Δrnt Δpnp rph-1 strains (data not shown), compared to the presence of primarily short poly(A) tails in the Δrnt rph-1 strain (Figure 1, lane 5). The increases in the level of all poly(A) tails in both triple mutants was consistent with the role played by PNPase and RNase II in regulating the in vivo poly(A) tail levels (54).
Nevertheless, in order to determine if the increased levels of poly(A) tails in the two triple mutants were specific to tRNAs, Northern analysis was carried out on the hisR and leuX tRNAs, the two tRNAs with the highest level of immature species in the Δrnt rph-1 double mutant (Table 2). Total RNA was pretreated with RNase H in the presence of oligo(dT20) to evaluate the length of the poly(A) tails. In the absence of PNPase, both hisR and leuX had identical percentages (data not shown) as well as similar lengths of immature species compared to the Δrnt rph-1 strain (Figure 9, lanes 1 and 5). However, in the absence of RNase II the hisR immature species were heterogeneous and the leuX immature species were slightly longer than in the Δrnt rph-1 strain (Figure 9, lanes 1 and 3). The longer leuX immature transcript in the absence of RNase II was consistent with its demonstrated role in the processing of the leuX downstream 3′ sequences (5). More importantly, treatment with RNase H did not change the length of immature species compared to the non-treated samples in any genetic background (Figure 9, lanes 2, 4 and 6). An identical Northern analysis of 5S rRNA showed a minor effect of both RNase II and PNPase (Figure 9, lanes 4 and 6) in shortening the poly(A) tails. However, the overall poly(A) tail lengths associated with 5S rRNA remained short.
Figure 9.
Northern analysis of hisR, leuX and 5S rRNA transcripts in RNase II and PNPase mutants after treatment with (+) or without (−) RNase H in the presence of oligo(dT20). Lanes 1 and 2, SK10148; lanes 3 and 4; SK10575; lanes 5 and 6, SK10609. Genotypes of each strain are noted above each lane. A star indicates immature and polyadenylated species. FL = full-length.
DISCUSSION
We have demonstrated here for the first time a role for polyadenylation in regulating the intracellular levels of functional tRNAs in E. coli. Essentially, RNase T and RNase PH, the two primary tRNA 3′ processing ribonucleases compete with PAP I for the 3′-ends of pre-tRNAs (Figure 10). Since the PAP I level in vivo is comparatively low in wild-type cells [30–50 molecules/cell (37)], RNase T and/or RNase PH rapidly process the majority of pre-tRNAs in wild-type cells to generate mature tRNAs (Figure 10), which are rapidly aminoacylated making them no longer substrates for PAP I. Increased polyadenylation of tRNA substrates in the presence of excess PAP I (Figure 8) is consistent with this model.
Figure 10.
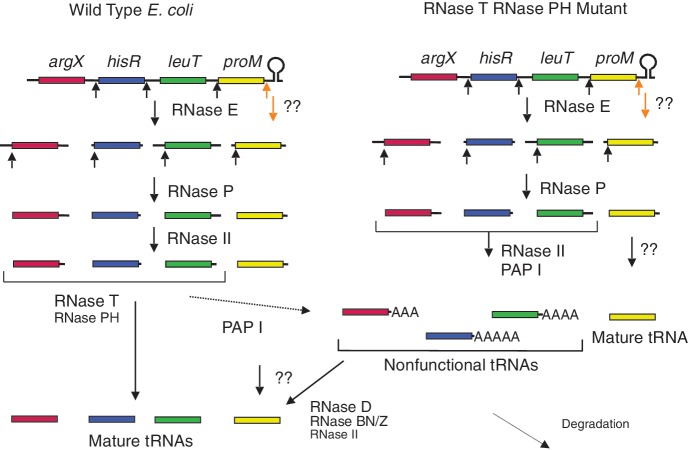
A schematic model for processing of the E. coli argX operon. RNase E cleaves the polycistronic transcript to generate the individual pre-tRNAs (1,2) followed by RNase P cleavages, which generate the mature 5′ ends on the pre-tRNAs. The mechanism for removal of the transcription terminator following proM is unknown (double question marks). The long 3′ trailer sequences associated with some pre-tRNAs generated by downstream RNase E cleavages are often trimmed back to 2–3 nt downstream of the CCA determinant by RNase II alone or by polyadenylation by PAP I and RNase II activity (10). In wild-type cells, the resulting argX, leuT and hisR pre-tRNAs are quickly processed at their 3′ ends by RNase T and/RNase PH. PAP I adds poly(A) tail to any unprocessed pre-tRNAs, which are very low in abundance in wild-type cells [(19), Figure 6]. The mechanism of 3′-end maturation of proM is not clear (double question marks). In the absence of either RNase T or RNase PH, the polyadenylated fraction of argX, leuT and hisR pre-tRNAs increases significantly (4,5). However, in the absence of both the ribonucleases majority of the argX, leuT and hisR pre-tRNAs are polyadenylated (Figure 6) resulting in a large increase of non-functional tRNAs (Table 4). Most of these tRNAs are very slowly processed (Table 3) to chargeable mature species probably by the action of RNase D, RNase BN/Z and RNase II (double question marks). Only a small fraction of the polyadenylated non-functional tRNAs are degraded, possibly by PNPase-dependent pathway (26).
In contrast, in the absence of both RNase T and RNase PH majority of the pre-tRNAs become substrates for polyadenylation by PAP I, generating significant levels of non-functional tRNAs (Figures 5 and 10; Table 4). Surprisingly, most of these species are processed very slowly (half-lives between 70 and 130 min, Table 3) presumably by the remaining tRNA processing enzymes such as RNase D, RNase BN/Z and RNase II (Figures 4 and 10). Only a small fraction of the polyadenylated species appear to be degraded via a general quality control process [Figures 4 and 10, (26)]. Of particular importance is the fact that the polyadenylation of pre-tRNAs occurs in wild-type cells (Figure 6), demonstrating that the competition among RNase T, RNase PH and PAP I is part of the normal tRNA processing pathway in E. coli.
Thus, the significant decreases in charged tRNA levels (Table 4) and increased cell generation time (Table 5) in the Δrnt rph-1 double mutant directly correlates with the 30-fold increase in poly(A) level (Figure 1 and Table 1). Furthermore, these defects are partially suppressed by inactivating PAP I (Tables 2–4). Although the activities of either RNase T or RNase PH appear to protect the vast majority of pre-tRNAs and mature tRNAs from PAP I mediated polyadenylation (Figure 3), the absence of RNase T alone leads to higher accumulation of short poly(A) tails (Figures 1 and 7) and significantly reduced growth rate (Table 2) compared to the absence of RNase PH. Taken together the above data indicated that RNase T was most effective in protecting the mature, stable RNA 3′-ends from polyadenylation.
Our data also show for the first time that poly(A) tail length in vivo is substrate dependent, indicating differential activity of PAP I on tRNAs and 5S rRNA versus other RNAs (mRNAs and 23S rRNA). For example, the poly(A) tails associated with tRNAs and 5S rRNA are predominantly short (≤5 nt) (Figures 1 and 6–9) compared to what has been observed for mRNAs (Figure 8B) (22,24,25,36,37), although poly(A) tails with an average length of 7–9 nt have been reported for the rpsO mRNA (55). These results raise the question as to why the poly(A) tails added onto tRNAs, 5S rRNA (Figures 6 and 7) and other stable RNAs (19) are inherently short compared to those found on mRNAs.
One obvious explanation is that the short poly(A) tails on stable RNAs arise from rapid shortening of longer tails by PNPase or RNase II as previously proposed (19). This model is quite attractive because several studies have shown that these major 3′–5′ exonucleases control the extent of polyadenylation of mRNAs (54,56,57). However, as shown in Figure 9, neither RNase II nor PNPase affected the length of the poly(A) tails associated with tRNAs and 5S rRNA. The data, however, are consistent with the requirement for ≥10 nt single-stranded regions for RNase II and PNPase to bind and degrade RNA substrates (56,58,59) as opposed to RNase T and RNase PH, which are very effective against single-stranded substrates up to 5–6 nt in length (60,61).
Moreover, it has been shown that Hfq, a RNA binding protein, affects PAP I processivity, both in vitro and in vivo (37,55,62). Interestingly, Hfq binds tRNAs with high specificity on its proximal surface, which is distinct from the poly(A) binding site (63). However, preliminary analysis comparing poly(A) tail profiles between Δrnt rph-1 and Δrnt rph-1 hfq-1 mutants did not reveal any significant difference in tRNA tail lengths (data not shown). It is possible that the short poly(A) tails found on tRNAs and 5S rRNA are simply due to a more distributive mode of PAP I activity on tRNAs compared to a more processive mode that is used for mRNA polyadenylation where Hfq plays a significant role.
Alternatively, the failure to elongate tRNA poly(A) tails, even after PAP I overexpression (Figure 8), could indicate a distinct polyadenylation reaction that may be similar to what is observed with the TRAMP complex in S. cerevisiae (64). This multiprotein complex contains a non-canonical PAP (Trf4p or Trf5p in S. cerevisiae, which is similar to E. coli PAP I), a Zn-knuckle protein (Air2P or Air1P) and an RNA helicase (Mtr4p/Dob1p) (64). Furthermore, the complex has been shown to target RNA substrates meant for further processing or exonucleolytic degradation by adding very short poly(A) tails (∼4–5 nt) (65). Of even more interest is the fact that recent data suggest that the RNA helicase component regulates the polyadenylation by the TRAMP complex (66). Since protein–protein interactions between PAP I and DEAD box RNA helicases in E. coli have already been demonstrated (67), it is possible that a similar mechanism for controlling the extent of the tRNA polyadenylation reaction exists in E. coli, but is not employed with mRNAs (37).
Although it has been shown that the addition of short poly(A) tails to defective pre-tRNAs can initiate their degradation (26) (Figure 10), the results presented in Figure 4 indicate that only a small fraction of the polyadenylated tRNA precursors are actually degraded. Rather, in the case of the hisR, cysT and leuX tRNAs, a significant portion of the polyadenylated tRNA precursors were actually converted into mature species with half-lives that ranged from 77 to 133 min (Table 3). This extremely slow conversion presumably is the result of very inefficient processing by some combination of RNase D, RNase BN/Z and perhaps RNase II (Figure 10). In comparison, 3′-end processing by RNase T and RNase PH is very rapid with half-lives so short that it is technically not possible to accurately measure them (Mohanty and Kushner, unpublished results). Taken together, our data suggest that the addition of poly(A) tails to pre-tRNAs and/or tRNAs by PAP I is primarily suppressed by both RNase T and RNase PH in wild-type cells.
The slow processing of the polyadenylated pre-tRNAs by the backup tRNA 3′-end maturation pathway that presumably utilizes RNase D, RNase BN/Z and RNase II (Figure 10) is somewhat surprising. However, it is interesting to note that except for RNase T, all of the other three 3′-tRNA processing enzymes, with the possible exception of RNase BN (13,68), can degrade through the encoded CCA terminus resulting in substrates that would not be repairable by tRNA nucleotidyl transferase (69). It is thus possible that the addition of short poly(A) tails to pre-tRNAs is designed to slow down 3′-end processing by the enzymes that could destroy tRNA functionality, improving the chances of maturation by RNase T. The unique substrate specificity of RNase T (60), the most effective among all the tRNA processing enzymes, is consistent with this hypothesis.
Surprisingly, a small number of tRNAs (7/50) were found not to be the substrates for either RNase T or RNase PH (Table 2). These tRNAs were also not substrates for PAP I even in a Δrnt rph-1 double mutant. Thus, no precursors of the proK, proL and proM tRNAs were observed in the absence of both RNase T and RNase PH (Figure 3, lanes 4 and 5). Even when PAP I was overexpressed these three tRNAs were not significantly polyadenylated (Mohanty and Kushner, manuscript in preparation). Although one can explain the lack of an RNase T and/or RNase PH effect based on the possibility that endonucleolytic cleavages occur immediately downstream of their CCA determinants, at this point it is not clear which endonuclease(s) might be carrying out this processing. Conversely, it is possible that these particular tRNAs are processed efficiently by exonucleases such as RNase D, RNase BN or RNase II.
On the other hand, it is more difficult to understand why only these seven tRNAs are not polyadenylated. All tRNAs have paired 5′- and 3′-ends, generally with a 4-nt single-stranded extension containing the CCA determinant, except for tRNAMetf1 and tRNAMetf2 (metY, metZ, metW and metV). All seven tRNAs resistant to polyadenylation have the same last 4-nt single-stranded extension (ACCA), but many of the tRNA species that are polyadenylated have the same 4-nt extension. However, there is one significant difference between the seven tRNAs that are not substrates for PAP I and the other 79 tRNA transcripts, namely a C nucleotide at the mature 5′-terminus. The vast majority of the tRNAs have G at their 5′-end with some having either an A (four species) or U (eight species) at this position. The presence of a 5′ C residue correlates perfectly with the 7/86 tRNAs that are resistant to polyadenylation, since the four asn tRNAs have an U in the 5′ position and they are subject to polyadenylation and one of the species with an A as the 5′-terminus (trpT) also gets polyadenylated (data not shown),
Finally, it has been demonstrated that overexpression of PAP I is toxic in a wild-type cell (24,70). Although the exact reason for this toxicity is still not well understood, the data presented in Figure 8, suggest that tRNAs are better PAP I substrates compared to mRNAs, considering the fact that after 15 min of PAP I induction >25% of the mature tRNAHis and tRNACys were polyadenylated (Figure 8, data not shown). This is in contrast to what has been observed with mRNAs under similar circumstances in which the lpp mRNA had the highest polyadenylation level of ∼5% (25). The concomitant increase in the level of mature tRNA polyadenylation (Figure 8) and decreased cell viability with increased time of PAP I induction (24) suggest a direct link between reduced levels of functional tRNAs, slower growth and eventual cell death (24). Taken together, it appears that polyadenylation in E. coli plays an important role in ensuring appropriate levels of functional tRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
Funding for open access charge: National Institutes of Health (grants GM57220 and GM81554 to S.R.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Westpheling for the generous gift of pSET152 plasmid DNA.
REFERENCES
- 1.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanty BK, Kushner SR. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007;35:7614–7625. doi: 10.1093/nar/gkm917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohanty BK, Kushner SR. Rho-independent transcription terminators inhibit RNase P processing of the secG leuU and metT tRNA polycistronic transcripts in Escherichia coli. Nucleic Acids Res. 2008;36:364–375. doi: 10.1093/nar/gkm991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohanty BK, Kushner SR. Processing of the Escherichia coli leuX tRNA transcript, encoding tRNAleu5, requires either the 3′−5′ exoribonuclease polynucleotide phosphorylase or RNase P to remove the Rho-independent transcription terminator. Nucleic Acids Res. 2010;38:5306–5318. doi: 10.1093/nar/gkp997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuven NB, Deutscher MP. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J. 1993;7:143–148. doi: 10.1096/fasebj.7.1.8422961. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic. Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Pandit S, Deutscher MP. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Deutscher MP. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem. 1994;269:6064–6071. [PubMed] [Google Scholar]
- 11.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 12.Dutta T, Deutscher MP. Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endoribonuclease. J. Biol. Chem. 2009;284:15425–15431. doi: 10.1074/jbc.M109.005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutta T, Deutscher MP. Mode of action of RNase BN/RNase Z on tRNA precursors: RNase BN does not remove the CCA sequence from tRNA. J. Biol. Chem. 2010;285:22874–22881. doi: 10.1074/jbc.M110.141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C, McClain WH, Altman S. Cleavage of tRNA precursors by the RNA subunit of E. coli ribonuclease P (M1 RNA) is influenced by 3′-proximal CCA in the substrates. Cell. 1984;38:219–224. doi: 10.1016/0092-8674(84)90543-9. [DOI] [PubMed] [Google Scholar]
- 15.King TC, Sirdeshmukh R, Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc. Natl Acad. Sci. USA. 1984;81:185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Deutscher MP. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl Acad. Sci. USA. 1995;92:6883–6886. doi: 10.1073/pnas.92.15.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidhardt FC, Umbarger HE. In: Escherichia coli and Salmonella. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Vol. 1. Washington, DC: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 18.Reuven NB, Deutscher MP. Substitution of the 3′ terminal adenosine residue of transfer RNA in vivo. Proc. Natl Acad. Sci. USA. 1993;90:4350–4353. doi: 10.1073/pnas.90.10.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. USA. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regnier P, Hajnsdorf E. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog. Mol. Biol. Transl. Sci. 2009;85:137–185. doi: 10.1016/S0079-6603(08)00804-0. [DOI] [PubMed] [Google Scholar]
- 21.Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. WIREs RNA. 2010;2:256–276. [Google Scholar]
- 22.O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohanty BK, Kushner SR. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol. Microbiol. 2002;45:1315–1324. doi: 10.1046/j.1365-2958.2002.03097.x. [DOI] [PubMed] [Google Scholar]
- 24.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 25.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen KG. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piedade J, Zilhao R, Arraiano CM. Construction and characterization of an absolute deletion of Escherichia coli ribonuclease II. FEMS Microbiol. Lett. 1995;127:187–193. doi: 10.1111/j.1574-6968.1995.tb07472.x. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100: 195–199. [PubMed] [Google Scholar]
- 32.Mohanty BK, Giladi H, Maples VF, Kushner SR. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol. 2008;447:3–29. doi: 10.1016/S0076-6879(08)02201-5. [DOI] [PubMed] [Google Scholar]
- 33.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 34.Ingle CA, Kushner SR. Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation. Proc. Natl Acad. Sci. USA. 1996;93:12926–12931. doi: 10.1073/pnas.93.23.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–146. doi: 10.1017/s1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ - 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 38.Dittmar KA, Mobley EM, Radek AJ, Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 2004;337:31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 39.Goodsell DS. Inside a living cell. Trends Biochem. Sci. 1991;16:203–206. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 40.Bremer H, Dennis PD. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt FC, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Vol. 2. Washington: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 41.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum E, Carpousis AJ, Higgins CF. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 43.Coburn GA, Mackie GA. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases is dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- 44.Mikkelsen ND, Gerdes K. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol. Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 45.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kruger MK, Sorensen MA. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 47.McClain WH, Jou YY, Bhattacharya S, Gabriel K, Schneider J. The reliability of in vivo structure-function analysis of tRNA aminoacylation. J. Mol. Biol. 1999;290:391–409. doi: 10.1006/jmbi.1999.2884. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen MA. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(-) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 2001;307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 49.Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Reports. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Misra TK, Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J. Biol. Chem. 1979;254:11154–11159. [PubMed] [Google Scholar]
- 51.Roy MK, Singh B, Ray BK, Apirion D. Maturation of 5S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur. J. Biochem. 1983;131:119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- 52.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 53.Murray EL, Schoenberg DR. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 2008;448:483–504. doi: 10.1016/S0076-6879(08)02624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohanty BK, Kushner SR. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- 55.Le Derout J, Folichon M, Briani F, Deho G, Regnier P, Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003;31:4017–4023. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Z-F, Deutscher MP. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 57.Folichon M, Allemand F, Regnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A) polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–463. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 58.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 59.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 60.Zuo Y, Deutscher MP. The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- 61.Kelly KO, Reuven NB, Li Z, Deutscher MP. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J. Biol. Chem. 1992;267:16015–16018. [PubMed] [Google Scholar]
- 62.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raynal LC, Carpousis AJ. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 68.Takaku H, Nashimoto M. Escherichia coli tRNase Z can shut down growth probably by removing amino acids from aminoacyl-tRNAs. Genes Cells. 2008;13:1087–1097. doi: 10.1111/j.1365-2443.2008.01230.x. [DOI] [PubMed] [Google Scholar]
- 69.Kelly KO, Deutscher MP. Characterization of Escherichia coli RNase PH. J. Biol. Chem. 1992;267:17153–17158. [PubMed] [Google Scholar]
- 70.Cao G-J, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl Acad. Sci. USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.