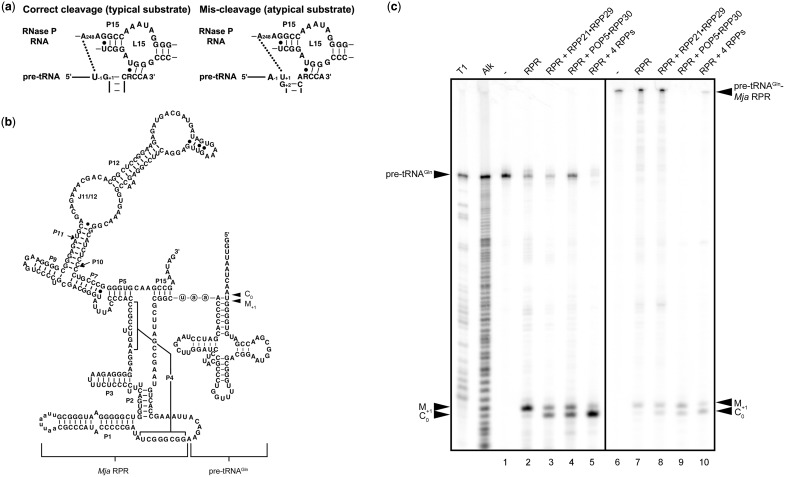
Figure 1.
Archaeal RPPs affect the RPR's fidelity of processing of a non-consensus substrate. (a) Possible interactions around the cleavage site between nucleotides in typical (consensus) or atypical (non-consensus) pre-tRNAs and the bacterial RPR. (b) Secondary structure of the self-cleaving pre-tRNAGln-Mja RPR used in this study. The site of tethering of pre-tRNAGln to Mja RPR is depicted. The 5′-aau-3′ spacer is shown in circled lowercase letters. The secondary structure follows conventional representations for all RPRs (100). (c) Rescue of RPR's mis-cleavage by the RPPs. T1, the G ladder generated by RNase T1 digestion of pre-tRNAGln. Alk, the ladder obtained from alkaline hydrolysis of pre-tRNAGln. Uncleaved pre-tRNAGln is shown in lane 1. Cleavage of pre-tRNAGln by Pfu RPR, RPR + RPP21•RPP29, RPR + POP5•RPP30 and RPR + both RPP complexes is shown in lanes 2–5, respectively. Lanes 6 and 7 represent uncleaved and self-cleaved pre-tRNAGln-Mja RPR, respectively. The cleaved 5′-leader products generated by addition of Mja RPP21•RPP29, POP5•RPP30 and four RPPs to pre-tRNAGln-Mja RPR are shown in lanes 8–10. M+1, the 5′-leader generated from mis-cleavage between positions +1 and +2 in pre-tRNAGln; C0, the 5′-leader from cleavage at the correct site between positions −1 and +1 in pre-tRNAGln.