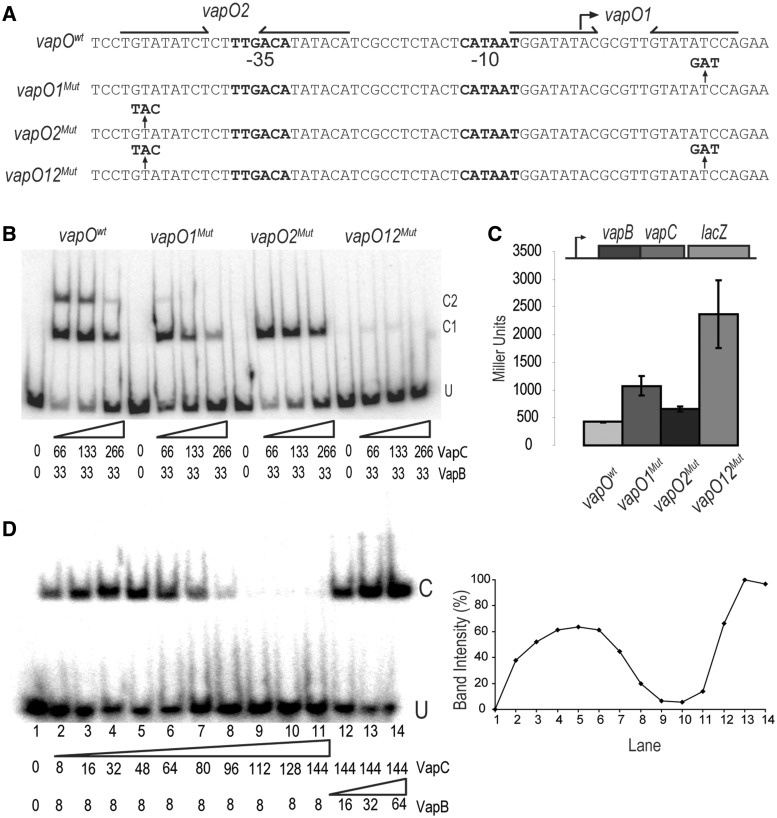
Figure 4.
One vapBC operator is sufficient for regulation by conditional cooperativity. (A) DNA sequences of the vapBC promoter region showing the base substitutions in vapO1Mut, vapO2Mut and vapO12Mut. Inverted repeats are indicated by arrows and promoter sequences by −10 and −35, respectively. (B) Gel shift assay of VapBC complex binding to the DNA fragments shown in (A). VapB and VapC were incubated with DNA (numbers are in nM) . U indicates unbound DNA fragment, C1 and C2 are fragments bound by either one or two VapBC complexes, respectively. (C) vapBC promoter activity in binding site mutants. TB28 (MG1655ΔlacIZYA) containing either Pwt::vapBCD7A::lacZYA (pKW512TFZD7A), P::vapO1Mut::vapBCD7A::lacZYA (pKW512TFZD7A-1), P::vapO2Mut::vapBCD7A::lacZYA (pKW512TFZD7A-2) or P::vapO1Mut::vapO2Mut::vapBCD7A::lacZYA (pKW512TFZD7A-1-2) were grown exponentially in LB medium at 37°C. At an OD600 of approximately 0.5, samples were collected and LacZ activity measured (Miller Units). (D) Gel shift analysis as in (B) but with a promoter DNA fragment (36 bp) containing only vapO1. Numbers below the gel are protein concentrations in nM (lane 1–14). Protein–DNA complexes were separated by 6% native PAGE. U and C indicate positions of unbound and bound DNA, respectively. Insert at right: Quantification of the C band-intensities (%) in the gel-shift shown at left.