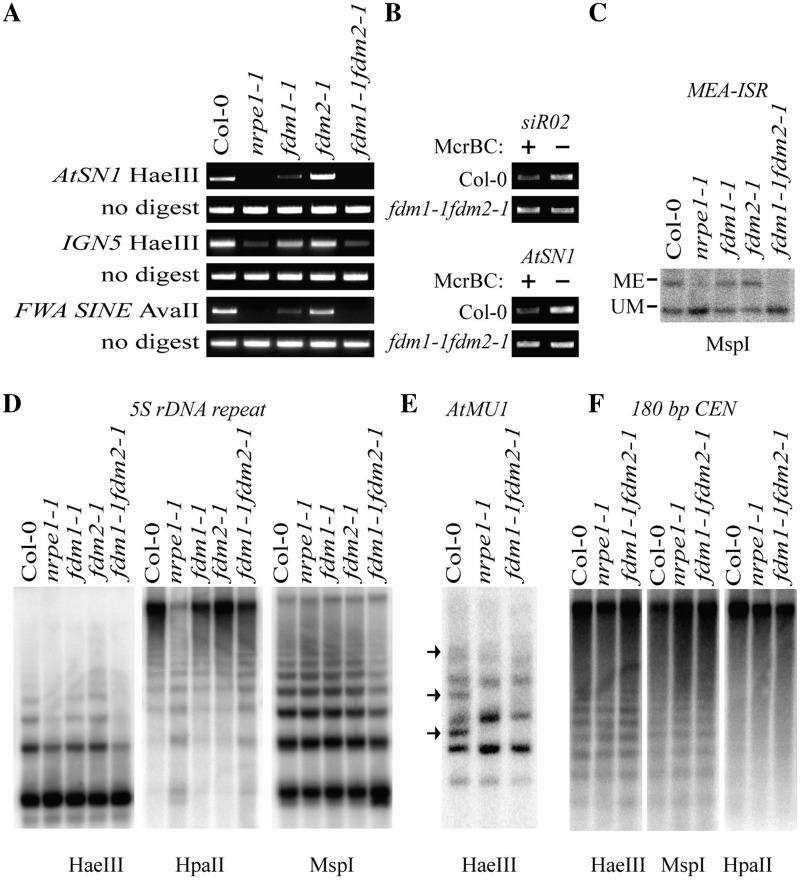
Figure 2.
FDM1 and FDM2 play redundant and essential roles in RdDM. (A) Reduced DNA methylation at AtSN1, IGN5 and FWA SINE in fdm1-1 fdm2-1. HaeIII-digested genomic DNAs from various genotypes were used for PCR amplification of AtSN1 and IGN5, whereas AvaII-treated genomic DNAs were used for the amplification of FWA SINE. Amplifications of undigested genomic DNA are used as loading controls. Col: WT plants. (B) Reduced DNA methylation at AtSN1 and siR02 loci in fdm1-1 fdm2-1. McrBC-digested and -undigested DNAs (control) were used for the amplification of AtSN1 and siR02. (C) Reduced DNA methylation at MEA-ISR in fdm1-1 fdm2-1. MspI-digested genomic DNAs from various genotypes were probed for MEA-ISR. Bands representing methylated (ME) or unmethylated (UM) DNA are indicated. (D) Reduced DNA methylation at 5S rDNA locus in fdm1-1 fdm2-1. HaeIII, HpaII or MspI-digested genomic DNAs from various genotypes were probed for 5S rDNA. (E) Reduced DNA methylation at AtMU1 locus in fdm1-1 fdm2-1. HaeIII-digested genomic DNAs were probed for AtMU1. The three undigested bands presented in Col (WT) but not in nrpe1-1 and fdm1-1 fdm2-1 were indicated by arrows. (F) Unaffected DNA methylation at 180 bp centromeric repeats in fdm1-1 fdm2-1. Following HpaII, MspI or HaeIII treatment, genomic DNAs from various genotypes were probed for 180-bp centromeric repeats.