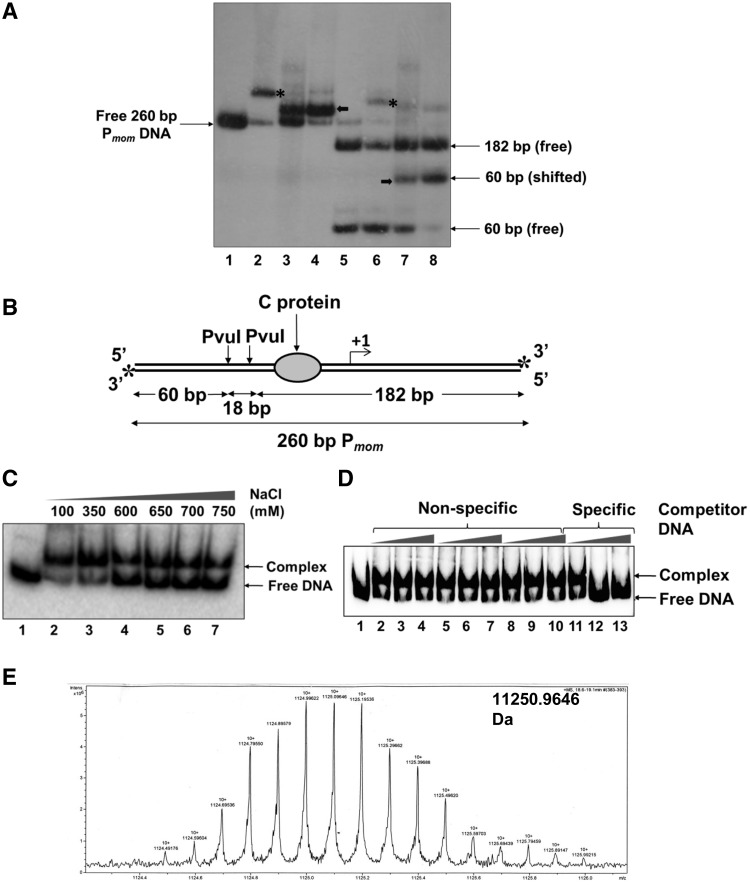
Figure 1.
Identification of the host factor that binds specifically to Pmom. (A) Pmom specific activity in crude cell extracts of E. coli. End-labeled 260-bp Pmom fragment (lane 1) was incubated with 220 nM purified C (lane 2) and 1 µg of crude sonicated extract (lane 3) and partially purified cell extract (lane 4) of E. coli MG1655. Lane 5 onwards, PvuI digested Pmom (lane 5) was incubated with 220 nM purified C (lane 6) and 1 µg of crude sonicated extract (lane 7) and partially purified cell extract (lane 8) of E. coli MG1655. EMSAs with cell extracts were carried out in the presence of 200 ng poly (dI – dC). Mobility shift caused by binding of C is depicted by asterisks while that caused by the unknown host factor is depicted by bold arrows. (B) Pmom fragment used for EMSAs. A 260-bp fragment of Pmom obtained upon digestion of plasmid pUW4 with EcoRI and HindIII was radiolabeled at both the 3′-ends (denoted by asterisks) by Klenow polymerase-mediated end filling. Digestion of this fragment with PvuI yields three fragments of sizes 60, 18 and 182 bp of which only the terminal fragments (60 and 182 bp) carry the radiolabel. The larger fragment (182 bp) contains the C-binding site whereas the smaller fragment (60 bp) comes from the upstream region. (C) Effect of ionic strength on stability of the complex. 220-bp Pmom obtained upon digestion of pUW4 with EcoRI and BamHI (lane 1) was incubated with 1 µg of crude cell extract of E. coli MG1655 in the presence of increasing concentrations of NaCl as indicated. The host factor–DNA complex was stable even in 750 mM NaCl. (D) Effect of non-specific competitor DNA on complex formation. End labeled 220 bp Pmom fragment (lane 1) was incubated with 1 µg of crude cell extract of E. coli MG1655 in the presence of 0-, 50- and 100-fold molar excess of three different unlabeled non-specific competitor DNA fragments (lanes 2–10) and unlabeled Pmom fragment (lanes 11–13). Labeled Pmom DNA is competed by the specific unlabeled Pmom DNA but not by any of the three non-specific DNA fragments, showing that the complex results from specific binding of a molecule in the extract to Pmom. (E) LC/ESI-MS analysis of the active fraction. Crude cell extract of E. coli MG1655 was processed as described in ‘Materials and Methods’ section. Eluted fractions from SP Sepharose column that tested positive for Pmom specific DNA binding activity in EMSA were pooled, dialyzed and subjected to LC/ESI–MS for whole protein molecular weight detection. Analysis of the fraction revealed a molecular mass of 11.25 kDa.