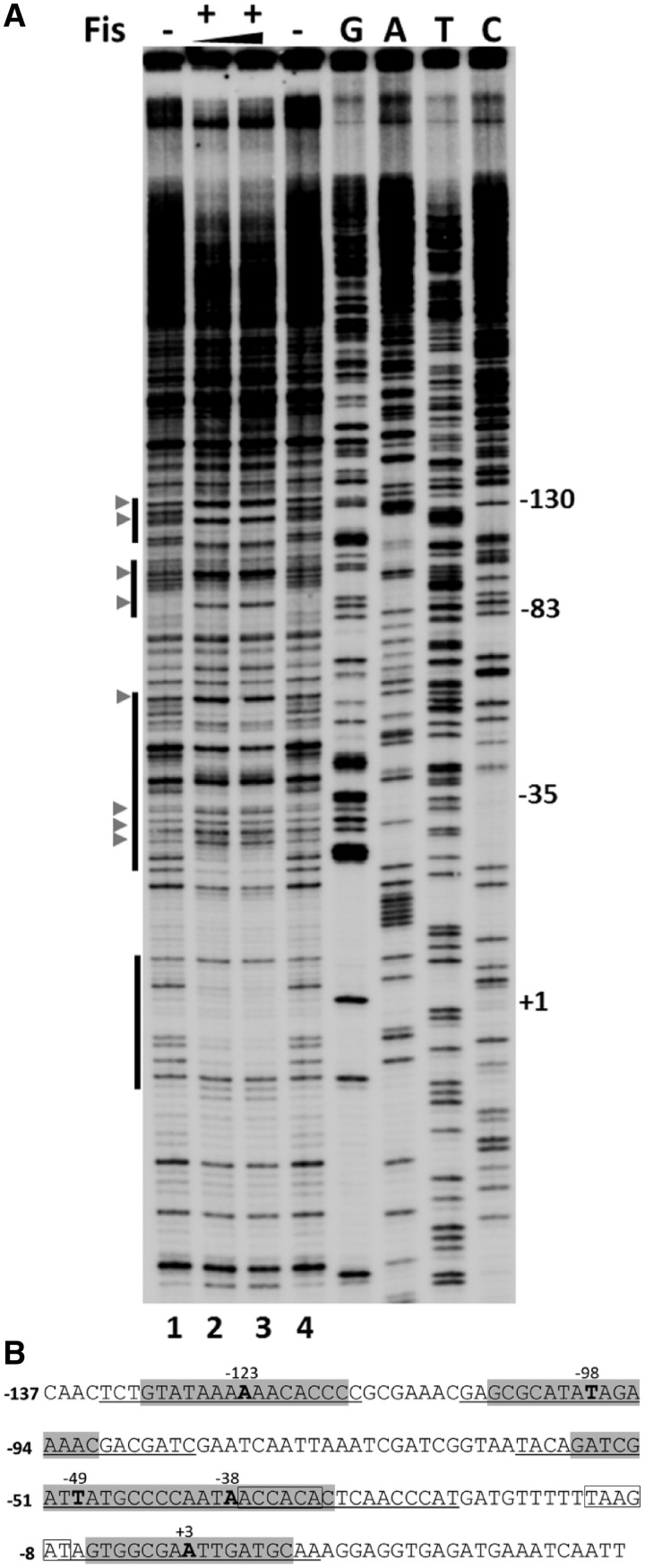
Figure 5.
Footprinting analysis of Fis binding at Pmom. (A) DNase I footprinting analysis of Fis binding sites in Pmom. Plasmid pUW4 was incubated with Fis (288 and 384 nM in lanes 2 and 3, respectively) prior to digestion with DNase I. Primer extension was carried out with Klenow polymerase using mom reverse primer. The vertical bars on the left side indicate regions protected from DNase I in the presence of Fis. G, A, T and C refer to Sanger's dideoxy sequencing ladders of Pmom obtained using mom reverse primer. Numbers on the right denote nucleotide positions with respect to the +1 start site of Pmom. DNase I hypersensitive sites are depicted by arrowheads. (B) Sequence of mom promoter. Regions protected by Fis are underlined and sequences matching with the consensus Fis binding site are highlighted with the central nucleotide emboldened and its position indicated. The −35 and −10 promoter elements are boxed.