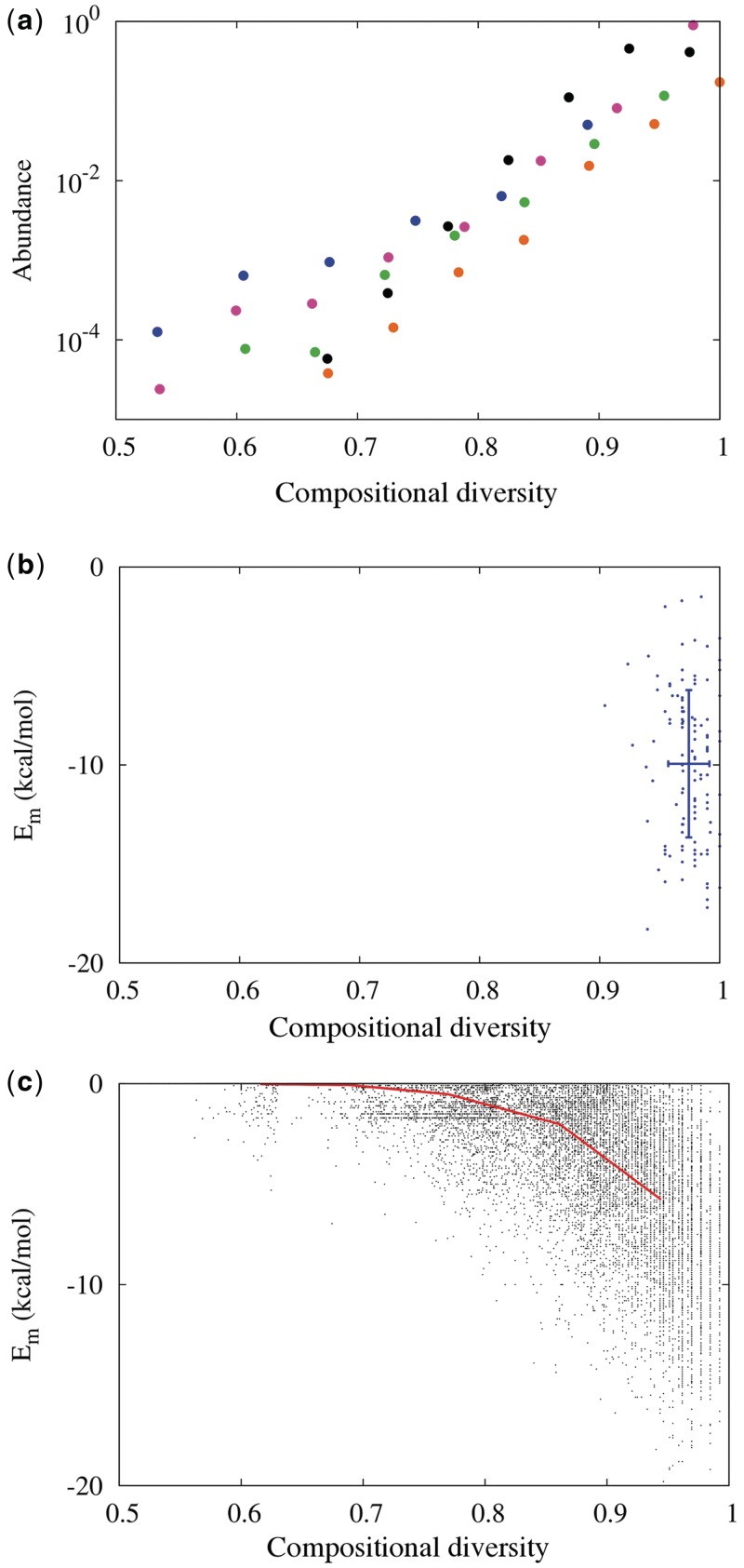
Figure 1.
Compositional diversity, sequence space and predicted RNA folding energy. (a) Most of sequence space is of high compositional diversity. Histogram of C4 for RNA sequences, computed from random sampling of 109 sequences of length 50 (black dots) in silico. The complete histogram for all possible sequences of shorter length is computable and is similar to that of the random sample of 50-mers (length 10 = blue, 12 = pink, 14 = green, 17 = orange). (b) Compositional diversity (C4) and predicted minimum folding energy (Em) for known ribozymes (length 40–60; see Supplementary Data) (45) are shown as blue dots with mean and SD (blue lines). (c) C4 versus Em (black dots) predicted by Viennafold (41) for 2.5 × 106 RNA sequences of length 50. To minimize effects from GC-content, we restricted the in silico sampling to sequences whose GC content is 40–60%. To avoid sampling artifacts, sequences were assigned to five bins according to C4, and an equal number of unique sequences were analyzed in each bin. The bin averages are shown as the red line (see Supplementary Data for values and SDs).