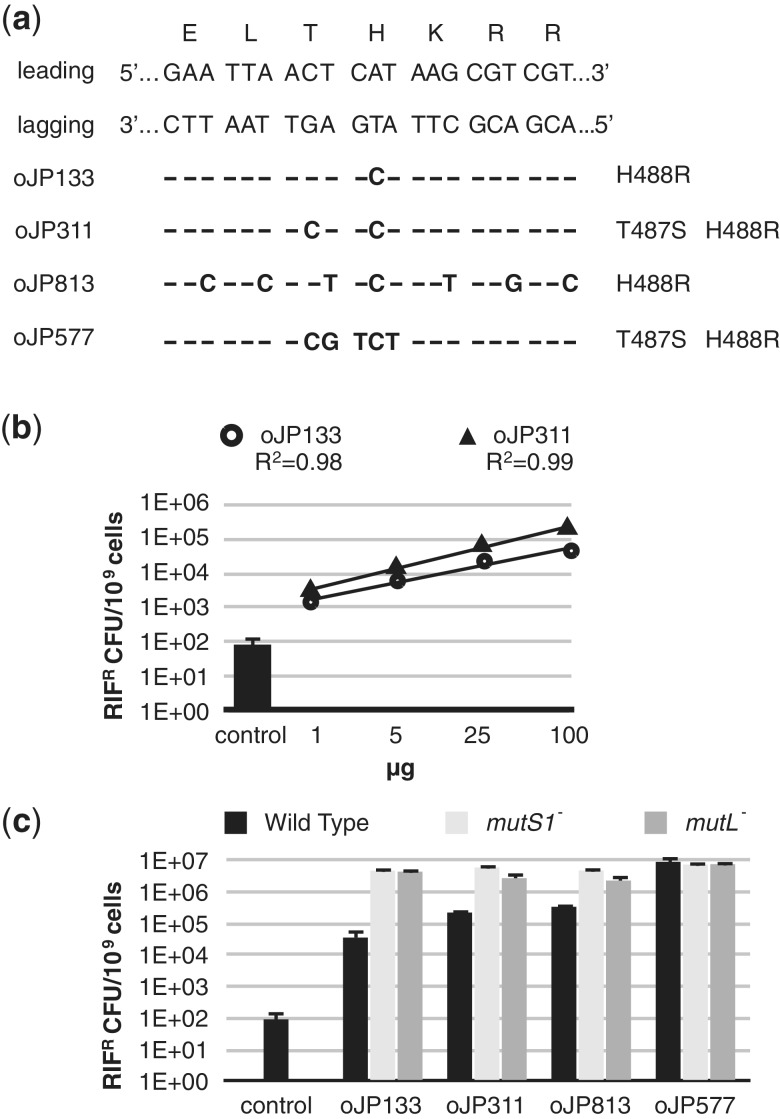
Figure 2.
Establishing and optimizing recombineering in L. reuteri. (a) The dsDNA sequence of the targeted rpoB region of L. reuteri is shown aligned with amino acids specified by each codon. On the left the leading and lagging strand are indicated and below the oligonucleotides are listed which result in different mismatches, all resulting in a rifampicin-resistant phenotype. The corresponding amino acid changes are listed on the right. (b) Titrations of the amount of oligonucleotides oJP133 (Ο) and oJP577 (Δ) were performed with 1, 5, 25 and 100 µg of oligonucleotide. Rifampicin resistant colonies derived from the control transformation are represented as a bar graph. Data are expressed as rifampicin-resistant cfu per 109 cells. Data shown are the averages of three independent experiments and error bars represent standard deviation. (c) Assessment of recombineering efficiencies with 100 µg oJP133, oJP311, oJP813 and oJP577 in the wild-type strain (black bars), and in the mismatch repair deficient strains MutS1− (light grey bars) and MutL− (dark grey bars). Data shown are the averages of three independent experiments and error bars represent standard deviation.