Figure 5.
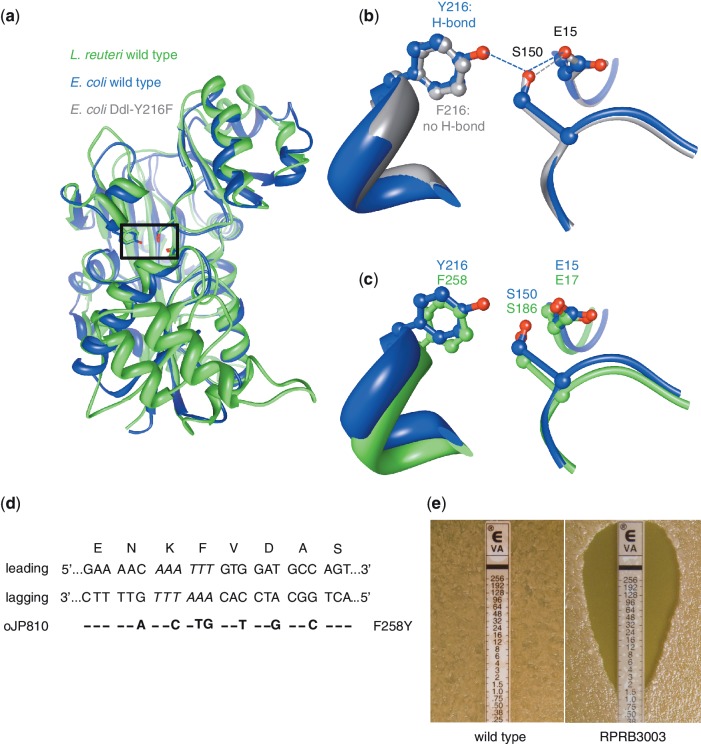
Converting vancomycin-resistant L. reuteri to vancomycin-sensitive by a single amino acid change. (a) The determined 3D structure of the d-ala-d-ala ligase (Ddl) protein of E.coli (PDB ID 1IOV; blue) and the modeled protein structure of Ddl of L. reuteri (green) were superimposed to locate the putative active site residues in Ddl of L. reuteri which, in E. coli, form a hydrogen-bonded network (Y216–E15–S150; boxed region). (b) Superimposing the determined structures of the Ddl proteins of the E. coli wild-type (blue) and a mutant (PDB ID 1IOW; grey) show that replacing a tyrosine at position 216 with a phenylalanine (Y216F) does not form a hydrogen-bond between F216–E15, and changes the enzymatic activity from a dipeptide ligase to a depsipeptide ligase. (c) We predicted the active site residues of Ddl-L. reuteri by superimposing the wild-type E. coli Ddl protein (blue) with the predicted protein structure of L. reuteri (green), and residues F258–E17–S186 in Ddl-L. reuteri correspond to the E. coli triad Y216–E15–S15, respectively. According to the E. coli model changing the phenylalanine at position 258 to a tyrosine (F258Y) in Ddl-L. reuteri would establish a hydrogen-bond between F258–E17 and may therefore yield dipeptide ligase activity which subsequently would result in a vancomycin-sensitive phenotype. (d) The dsDNA sequence of the ddl region of L. reuteri is shown aligned with the encoded amino acids for each triplet. Italic sequence represents the ApoI restriction site that is mutated by incorporation of oJP810. On the left are the leading and lagging strand indicated, and below is oJP810 showing the different mismatches with the resulting amino acid change (F258Y) on the right. (e) Susceptibility of L. reuteri wild-type (left) and the mutant RPRB3003 (right) to vancomycin using an Etest assay. The MIC is determined by identifying where bacterial growth intersects the Etest strip.