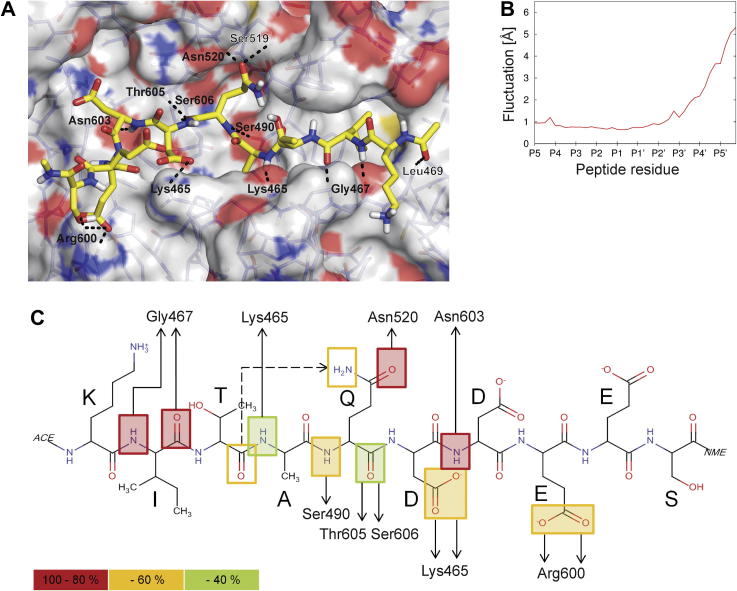
Fig. 5.
Molecular Dynamics simulation of the Plasmodium falciparum subtilisin-like protease 1 (PfSUB1) structure in complex with SERA4st1. (A) Binding mode of SERA4st1 extracted after 40 ns from the MD simulation. Hydrogen bond interactions between the peptide (yellow carbon coloured stick) and PfSUB1 are indicated by dashed lines. (B) Root-mean square fluctuations (RMSF) about the mean position of the peptide backbone atoms. For RMSF calculation, overall translational and rotational motions were removed with respect to the Cα atoms of the PfSUB1 structure. (C) Scheme of hydrogen bonds formed during the MD simulation between PfSUB1 and peptide backbone and side-chain atoms. The peptide orientation is shown from N- to C-terminus left-to-right, and thus is opposite to the orientation shown in (A). Very strong hydrogen bonds (occupancy of 80–100%) are boxed in red, strong hydrogen bonds (occupancy of 60–80%) are in orange, and weaker hydrogen bonds (occupancy of 40–60%) are shown in green. Occupancy values were obtained over the course of the entire 50 ns of simulation time. Repeating the analysis for the first and second half of the trajectory resulted in the same hydrogen bond pattern.