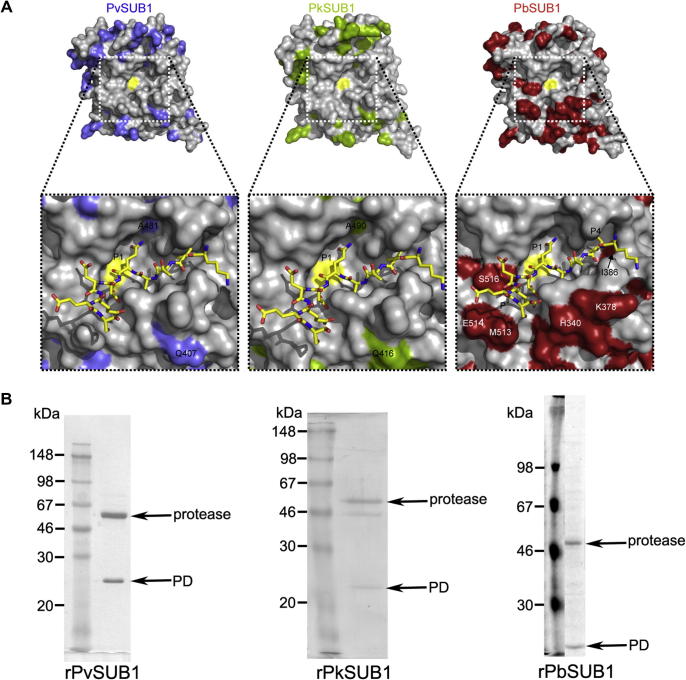
Fig. 7.
Homology modelling and recombinant expression reveals similarities and differences between Plasmodium subtilisin-like protease 1 (SUB1) orthologues. (A) Molecular surface representation of homology models of the Plasmodium vivax (Pv)SUB1, Plasmodium knowlesi (Pk)SUB1 and Plasmodium berghei (Pb)SUB1 catalytic domains. Coloured residues (blue for PvSUB1, green for PkSUB1, red for PbSUB1) indicate positions that are different from structurally equivalent residues in PfSUB1, whereas identical residues are shown in grey. The catalytic Ser is shown in yellow. White dashed frames delimitate the active site grooves, and the zoomed views below show the modelled binding mode of SERA4st1 (KITAQ ↓ DDEES). PvSUB1 and PkSUB1 are almost identical to PfSUB1 in the active site region, with the only two substitutions being Ala481 and Gln407 in PvSUB1, and Ala490 and Gln416 in PkSUB1. In contrast, PbSUB1 diverges significantly, with six substitutions (His340, Lys378, Ile386, Met513, Glu514 and Ser516), indicating likely differences in the specificity of this enzyme at the P4 and P′ positions. (B) Recombinant expression of SUB1 orthologues. Proteins were expressed using baculovirus-infected insect cells and purified as described in Section 2.1. Positions of migration of the protease domain and the prodomain species resulting from cleavage during maturation are indicated. The left-hand track on each gel contains molecular mass standards of the indicated sizes. Predicted molecular masses of the protease domains resulting from cleavage at the expected autocatalytic processing site (Supplementary Fig. S1) are: PvSUB1, 48,082 Da; PkSUB1, 49,072 Da; and PbSUB1, 44,868 Da.