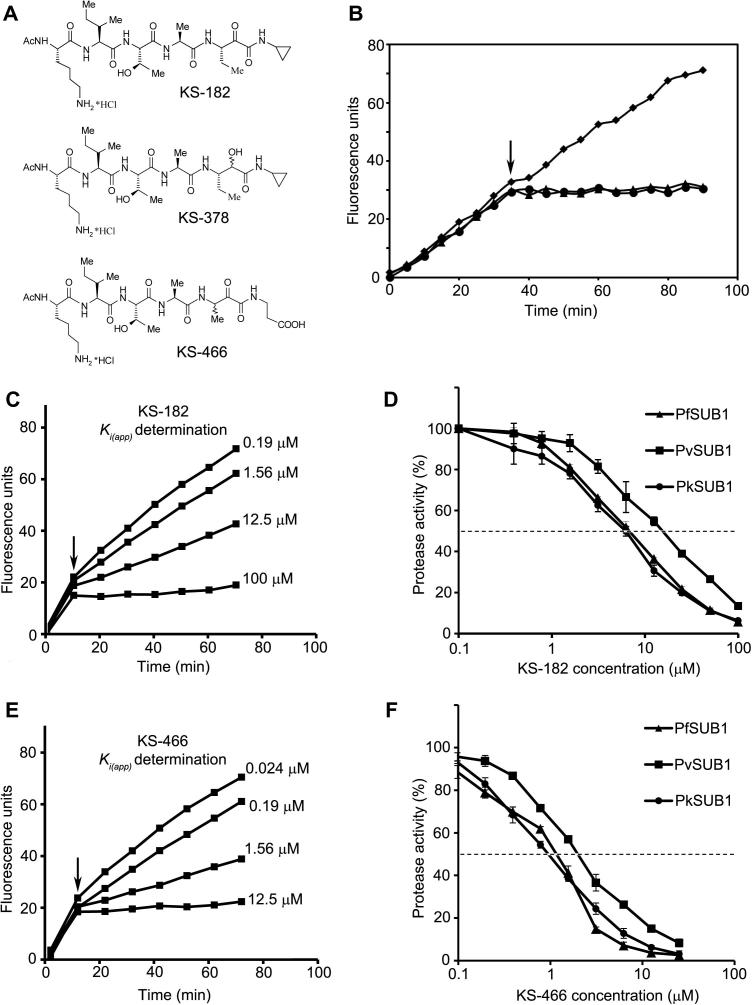
Fig. 9.
Peptidyl alpha-ketoamides based on a Plasmodium falciparum subtilisin-like protease 1 (PfSUB1) substrate are micromolar inhibitors of PfSUB1, Plasmodium vivax SUB1 (PvSUB1) and Plasmodium knowlesi SUB1 (PkSUB1). (A) Structures of the alpha-ketoamide KS-182, the corresponding aminoalcohol KS-378, and the extended KS-182 derivative KS-466. (B) Typical progress curve showing the profile of fluorescence increase with time produced by cleavage of substrate SERA4st1F-6R12 (0.1 μM) by recombinant P. falciparum SUB1 catalytic domain (rPfSUB1cat; 1 U/ml). The arrow indicates the point at which the reactions were supplemented with KS-182 (100 μM final, added from a 10 mM stock solution in DMSO; closed triangles), 1 mM para-hydroxymercuribenzoate (positive control inhibitor, closed circles), or 1% v/v DMSO (vehicle only control, closed squares). (C) and (E) Determination of Ki(app) values for inhibition of rPfSUB1cat by KS-182 and KS-466. Progress curves showing the effects of adding different concentrations of the peptidyl alpha-ketoamides to an ongoing digestion are as in A. The point of compound addition is indicated by an arrow in each plot. In all cases, compounds were diluted 1:100 into the digestion reactions, from stock dilutions in DMSO. The final DMSO concentration in each reaction was therefore invariant at 1% (v/v). Values of Ki(app) were calculated as described in Section 2.3. Since the concentration of substrate SERA4st1F-6R12 used for these experiments (0.1 μM) is well below its km, the calculated Ki(app) is unlikely to be significantly different from the Ki (Nicklin and Barrett, 1984). (D and F) Dose–response curves showing the relationship between concentrations of KS-182 or KS-466, and inhibition of rPfSUB1cat, rPkSUB1cat, and rPvSUB1cat. Calculated IC50 values are 6 μM, 6 μM and 12 μM, respectively, for KS-182, or 1 μm, 1 μM and 2 μM for KS-466. Points are means of triplicate measurements and error bars indicate S.D. values. The horizontal dotted line in each plot indicates 50% inhibition of protease activity.