Abstract
A single nucleotide polymorphism (SNP) rs999737 at 14q24.1 was identified as a susceptibility marker of breast cancer in a genome-wide association study of the European population, which was also confirmed by some of the following studies in populations of European descent. However, rs999737 is very rare or nonpolymorphic in non-Europeans including Chinese, and the role of other genetic variants at 14q24.1 has not been evaluated in populations of non-European descent. In this study, we first selected 21 common tagging SNPs (minor allele frequency [MAF] >0.05 in the Chinese population) by searching the Hapmap database, covering a linage disequilibrium region of more than 70 Kb at 14q24.1, and then conducted a two-stage study (stage I: 878 cases and 900 controls; stage II: 914 cases and 967 controls) to investigate the associations between these tagging SNPs and risk of breast cancer in a Chinese population. In stage I, two SNPs (rs2842346 and rs17828907) were identified to be significantly associated with breast cancer risk (p=0.030 and 0.027 for genotype distributions, respectively). However, no significant associations were found between these two SNPs and breast cancer risk in either stage II or the combined dataset. These findings suggest that common variants at 14q24.1 might not be associated with the risk of breast cancer in the Chinese population, which will need the replication in additional larger studies.
Introduction
Breast cancer is the most common malignancy among women around the world. The incidence has been increasing in recent two decades in China, reaching 24.8 per 100,000 in 2005 as estimated (Yang et al., 2005). In addition to unfavorable environmental exposures and lifestyles, genetic factors have been implicated in the susceptibility to breast cancer (Lichtenstein et al., 2000). However, approaches including a family-based linkage study and an association study based on candidate genes have been in a dilemma in identifying common genetic variants that confer the susceptibility to complex diseases including breast cancer (Breast Cancer Association Consortium, 2006), because few could be confirmed by the following studies. Recently, genome-wide association studies (GWAS) have been successful in identifying susceptibility loci for many diseases (Manolio, 2010), and more than one dozen of single nucleotide polymorphisms (SNPs) have been found for breast cancer (Turnbull et al., 2010). However, most GWAS were performed in populations of European descent, and, thus, it is important to evaluate whether these associations could be applicable to diverse populations across different descents.
In 2009, Thomas et al. conducted a three-stage GWAS with a total of 9,770 cases and 10,799 controls and reported an SNP rs999737 at 14q24.1 as a novel susceptibility marker for breast cancer in the European population. Later, another GWAS and a combined analysis of GWASs (Turnbull et al., 2010; Li et al., 2011) also confirmed the association between rs999737 and breast cancer risk in Europeans. As shown in Figure 1, this SNP maps to a more than 70-Kb linkage disequilibrium (LD) block entirely contained within intron 12 of RAD51L1 gene, which is involved in the key reactions of a homologous recombination by interacting with other paralogs in the RAD51 protein family (Miller et al., 2002; Lio et al., 2003). However, rs999737 is very rare or nonpolymorphic in other populations [Chinese (MAF: 0.011), Japanese (MAF: 0), and Africans (MAF: 0)], based on the HapMap database. Thus, this SNP does not appear to be an important marker for breast cancer risk in populations of non-European descent. Considering the implication of 14q24.1 region in breast cancer risk identified by GWASs, we hypothesize that other genetic variants at 14q24.1 may be associated with the risk of breast cancer and serve as susceptibility markers for non-European populations. Therefore, we refined this region and conducted a two-stage case-control study with 1792 breast cancer cases and 1867 controls to investigate the association between 21 tagging SNPs at 14q24.1 and breast cancer risk in a Chinese population.
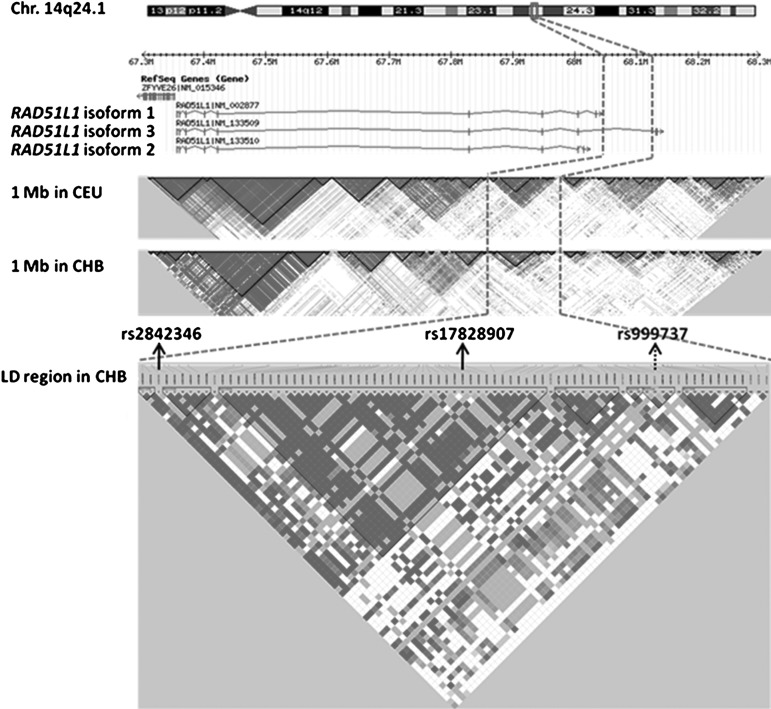
FIG. 1.
Linkage equilibrium (LD) plots and gene locations in the region of chromosome 14q24.1. The LD patterns were shown for the region of 67.3–68.3 Mb around the SNP rs999737 (indicated by the dashed arrow) in European (CEU) and Chinese (CHB) populations based on HapMap data (Rel 24/phase II Nov08). The LD region including rs999737 (MAF=0.011 in CHB) was restricted to chr14:68044030–68121653 by two recombination hot spots and was entirely mapped to the intron 12 of the RAD51L1 encoding the longest transcript (isoform 3). The SNPs rs2842346 and rs17828907 in the LD region of CHB were significantly associated with breast cancer risk in stage I, but not validated in stage II. SNP, single nucleotide polymorphism.
Materials and Methods
Study subjects
In this study, breast cancer cases were histopathologically diagnosed and consecutively recruited from the First Affiliated Hospital of Nanjing Medical University, the Cancer Hospital of Jiangsu Province, and the Gulou Hospital, Nanjing, China, between January 2004 and April 2010, which had been previously described (Ma et al., 2006). Those who reported previous cancer history, metastasized cancer from other organs, or previous radiotherapy or chemotherapy were excluded. All cancer-free women (controls) were from a cohort of more than 30,000 participants at the same period as the patients were recruited, which was established in 2004 and 2005 with the aim of exploring the environmental and genetic risk factors for common chronic diseases in the Wujin county of Changzhou City, Jiangsu Province, China. Based on the frequency matching of age (±5 years), 1867 controls were randomly selected from all women subjects in this cohort by the simple random sampling. All participants were genetically unrelated, ethnic Han Chinese women. After signing informed consent, each woman was interviewed face-to-face by trained interviewers using a structure questionnaire including information on demographic characteristics, menstrual and reproductive history, and environmental exposure. Each subject was required to donate 5 mL of venous blood after the interview. A total of 878 breast cancer cases and 900 controls were selected for the first screening stage, and the remaining 914 cases and 967 controls were further genotyped for the validation stage. The information of estrogen receptor (ER) and progesterone receptor (PR) status determined by immunohistochemistry examinations were obtained from medical records of patients in the hospitals. This study was approved by the institutional review board of Nanjing Medical University.
SNPs selection and genotyping
The LD region (chr14:68044030–68121653) including rs999737 was separated by two recombination spots at the position of chromosome 14q24.1 (Fig. 1). In this region, 161 SNPs were genotyped in the Chinese Han population (CHB) in HapMap (Rel 24/phase II Nov 08). A pair-wise approach with an r2 threshold of 0.80 was applied to define the tagging SNPs for common SNPs with MAF >0.05. As shown in Table 2, 21 loci were finally selected as tagging SNPs to cover this LD region in the CHB population.
Table 2.
Genotyping Results of Tagging SNPs at 14q24.1 and Distributions among Cases and Controls
| |
|
|
|
|
Minor allele frequency |
|
|
|---|---|---|---|---|---|---|---|
| SNP | Position | Alleles | Call rate | HWEa | Case | Control | pb |
| rs2842346 | 68045127 | C>T | 0.989 | 0.879 | 0.155 | 0.125 | 0.030 |
| rs2842344 | 68046724 | T>C | 0.988 | 0.859 | 0.091 | 0.105 | 0.335 |
| rs10131789 | 68047253 | C>T | 0.974 | 0.458 | 0.463 | 0.473 | 0.788 |
| rs2842341 | 68048542 | G>A | 0.983 | 1.000 | 0.267 | 0.268 | 0.951 |
| rs2525518 | 68068686 | G>A | 0.980 | 0.696 | 0.094 | 0.095 | 0.972 |
| rs2189517 | 68072741 | G>A | 0.985 | 0.408 | 0.289 | 0.283 | 0.912 |
| rs12882030 | 68075036 | A>G | 0.974 | 0.148 | 0.365 | 0.370 | 0.952 |
| rs17828907 | 68080958 | A>T | 0.990 | 0.298 | 0.073 | 0.089 | 0.027 |
| rs12587232 | 68097619 | A>G | 0.988 | 0.686 | 0.471 | 0.471 | 0.988 |
| rs17105837 | 68098494 | G>A | 0.979 | 0.719 | 0.394 | 0.375 | 0.493 |
| rs2525504 | 68098726 | A>G | 0.942 | 0.771 | 0.370 | 0.376 | 0.587 |
| rs11158751 | 68101178 | G>A | 0.987 | 0.873 | 0.292 | 0.299 | 0.542 |
| rs1468280 | 68101613 | A>G | 0.971 | 0.144 | 0.405 | 0.415 | 0.312 |
| rs8007194 | 68105178 | G>A | 0.994 | 0.879 | 0.111 | 0.124 | 0.425 |
| rs6573841 | 68107274 | C>T | 0.984 | 0.837 | 0.088 | 0.088 | 0.632 |
| rs6573842 | 68107690 | T>C | 0.989 | 0.420 | 0.209 | 0.211 | 0.799 |
| rs11621880 | 68108799 | C>T | 0.988 | 0.575 | 0.241 | 0.234 | 0.822 |
| rs2285883 | 68113723 | G>A | 0.989 | 0.692 | 0.213 | 0.217 | 0.837 |
| rs1290999 | 68115629 | A>G | 0.984 | 0.349 | 0.300 | 0.313 | 0.498 |
| rs2253168 | 68120218 | A>G | 0.970 | 0.586 | 0.437 | 0.446 | 0.306 |
| rs761944 | 68121653 | T>G | 0.987 | 0.175 | 0.356 | 0.374 | 0.321 |
p-values for Hardy–Weinberg equilibrium tests.
Derived from χ2 tests for distribution of genotypes between cases and controls.
SNP, single nucleotide polymorphism.
In stage I, genotyping was performed by using the TaqMan OpenArray Genotyping System (Life Technologies, Carlsbad, CA), a medium-throughput genotyping platform. Normalized DNA samples were loaded and amplified on arrays as recommended by the manufacturer. Two blank controls were contained in each 48-sample array. For the endpoint assay, arrays were scanned on the OpenArray NT imager, and genotypes were called using the OpenArray SNP Genotyping analysis software. The call rates ranged from 94.2% to 99.4% for 21 SNPs genotyped in stage I, and the mean call rate reached 98.2%. In stage II, the significant SNPs identified in stage 1 were further genotyped in 914 breast cancer cases and 967 controls by using the TaqMan assay based on the ABI 7900 System (Life Technologies). Genotyping was performed without knowing the subjects' case or control status, and two NTCs in each 384-well format were used for quality control. The accordance achieved 100% for the duplicates of 10% of samples.
Statistical analyses
The χ2 tests or Student's t-tests were used to evaluate the difference of distributions among cases and controls for categorical or continuous variables, respectively. Logistic regression analyses were employed to detect the associations between SNPs and the risk of breast cancer by estimating odds ratios (ORs) and their 95% confidence intervals (CIs). The adjustment factors for genetic associations included age, age at menarche, and menopausal status. Goodness-of-fit χ2 tests were performed to test Hardy–Weinberg equilibrium by comparing the observed genotype frequencies with the expected ones among the controls. The heterogeneity between subgroups was assessed with the χ2-based Q test, and the heterogeneity was considered significant when p<0.10. All the statistical analyses were performed with Statistical Analysis System software (9.1.3; SAS Institute, Cary, NC).
Results
The distributions of selected variables between cases and controls have been described elsewhere (Han et al., 2011). Briefly, the age between the cases and controls was comparable (p>0.05), but breast cancer cases had an earlier age at menarche and a later age at first live birth when compared with controls (p<0.001) (Table 1). The proportion of nature menopausal age ≥50 (62.8%) was higher in cases than in controls (52.3%) (p=0.018). There were 803 (55.5%) cases with ER of positive status and 810 (56.1%) cases with PR of positive status.
Table 1.
Distribution of Selected Variables between Breast Cancer Cases and Controls
| |
Stage I |
Stage II |
Combined dataset |
|
|||
|---|---|---|---|---|---|---|---|
| Variables | Cases n=878 (%) | Controls n=900 (%) | Cases n=914 (%) | Controls n=967 (%) | Cases n=1792 (%) | Controls n=1867 (%) | Combined pa |
| Age at diagnosis or recruitment (year) | |||||||
| Mean±SD | 51.29±11.38 | 51.47±11.67 | 50.11±11.36 | 48.64±12.28 | 50.69±11.38 | 50.01±12.07 | 0.077 |
| < 50 | 469 (53.4) | 472 (52.4) | 516 (56.5) | 548 (56.7) | 985 (55.0) | 1020 (54.6) | |
| ≥50 | 409 (46.6) | 428 (47.6) | 398 (43.5) | 419 (43.3) | 807 (45.0) | 847 (45.4) | |
| Age at menarche (year) | |||||||
| Mean±SD | 15.25±1.8 | 15.85±1.9 | 15.18±1.96 | 16.24±1.81 | 15.21±1.90 | 16.05±1.86 | <0.0001 |
| <15 | 501 (58.1) | 395 (44.1) | 528 (59.1) | 340 (35.3) | 1029 (58.6) | 735 (39.6) | |
| ≥15 | 361 (41.9) | 501 (55.9) | 365 (40.9) | 622 (64.7) | 726 (41.4) | 1123 (60.4) | |
| Missing | 16 | 4 | 21 | 5 | 37 | 9 | |
| Age at first live birth (year) | |||||||
| Mean±SD | 25.65±3.48 | 24.90±3.40 | 25.55±3.16 | 24.15±2.58 | 25.60±3.32 | 24.51±3.03 | <0.0001 |
| < 25 | 432 (52.0) | 547 (62.7) | 429 (50.5) | 675 (71.6) | 861 (51.3) | 1222 (67.3) | |
| ≥25 | 398 (48.0) | 325 (37.3) | 421 (49.5) | 268 (28.4) | 819 (48.7) | 593 (32.7) | |
| Missing | 48 | 28 | 64 | 24 | 112 | 52 | |
| Menopausal status | |||||||
| Premenopausal | 416 (48.4) | 437 (49.4) | 434 (48.4) | 530 (55.4) | 850 (48.4) | 967 (52.4) | <0.0001 |
| Postmenopausal | 386 (44.9) | 432 (48.8) | 379 (42.3) | 418 (43.7) | 765 (43.5) | 850 (46.1) | |
| Unnatural menopause | 58 (6.7) | 16 (1.8) | 84 (9.3) | 9 (0.9) | 142 (8.1) | 25 (1.5) | |
| Missing | 18 | 15 | 17 | 10 | 35 | 25 | |
| Age at natural menopause (year) | |||||||
| Mean±SD | 49.85±3.18 | 49.15±4.09 | 49.71±3.72 | 49.69±3.56 | 49.78±3.45 | 49.42±3.84 | 0.018 |
| < 50 | 145 (38.2) | 207 (50.4) | 131 (36.1) | 179 (45.0) | 276 (37.2) | 386 (47.7) | |
| ≥50 | 235 (61.8) | 204 (49.6) | 232 (63.9) | 219 (55.0) | 467 (62.8) | 423 (52.3) | |
| Missing | 6 | 21 | 16 | 20 | 22 | 41 | |
| Estrogen receptor (ER) | |||||||
| Negative | 321 (46.5) | 322 (42.6) | 643 (44.5) | ||||
| Positive | 369 (53.5) | 434 (57.4) | 803 (55.5) | ||||
| Missing | 188 | 158 | 346 | ||||
| Progesterone receptor (PR) | |||||||
| Negative | 294 (42.6) | 340 (45.1) | 634 (43.9) | ||||
| Positive | 396 (57.4) | 414 (54.9) | 810 (56.1) | ||||
| Missing | 188 | 160 | 348 | ||||
t-tests and χ2 tests were used for continuous or categorical variables, respectively.
In stage I, genotypes of all 21 SNPs among controls were consistent with the Hardy–Weinberg equilibrium (p>0.05). Among the 21 loci, only the genotype distributions of two SNPs were significantly different between cases and controls (p=0.030 for rs2842346 and p=0.027 for rs17828907, respectively) (Table 2). Logistic regression analyses revealed that the variant genotypes of rs2842346 were significantly associated with an increased risk of breast cancer (CT/TT vs. CC: adjusted OR=1.27, 95% CI=1.02–1.58; additive model: adjusted OR=1.26, 95% CI=1.03–1.53), while the variant genotypes of rs17828907 showed a protective effect on breast cancer risk (AT/TT vs. AA: adjusted OR=0.74, 95% CI=0.57–0.97) (Table 3).
Table 3.
The Association between Rs2842346 and Rs17828907 and Risk of Breast Cancer
| |
Stage I |
Stage II |
Combined dataset |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Cases n (%) | Controls n (%) | OR (95% CI)a | pa | Cases n (%) | Controls n (%) | OR (95% CI)a | pa | Cases n (%) | Controls n (%) | OR (95% CI)a | pa |
| rs2842346 | 860 | 899 | 902 | 955 | 1762 | 1854 | ||||||
| CC | 613 (71.3) | 688 (76.5) | 1.00 | 685 (75.9) | 726 (76.0) | 1.00 | 1298 (73.7) | 1414 (76.3) | 1.00 | |||
| CT | 227 (26.4) | 198 (22.0) | 1.25 (0.99–1.56) | 0.859 | 200 (22.2) | 218 (22.8) | 0.92 (0.73–1.16) | 0.176 | 427 (24.2) | 416 (22.4) | 1.06 (0.91–1.25) | 0.242 |
| TT | 20 (2.3) | 13 (1.5) | 1.67 (0.82–3.42) | 0.270 | 17 (1.9) | 11 (1.2) | 1.59 (0.71–3.59) | 0.222 | 37 (2.1) | 24 (1.3) | 1.63 (0.95–2.78) | 0.098 |
| CT+TT | 247 (28.7) | 211 (23.5) | 1.27 (1.02–1.58) | 0.031 | 217 (24.1) | 229 (24.0) | 0.95 (0.76–1.20) | 0.684 | 464 (26.3) | 440 (23.7) | 1.09 (0.94–1.28) | 0.261 |
| Additive model | — | — | 1.26 (1.03–1.53) | 0.023 | — | — | 0.99 (0.81–1.22) | 0.949 | — | — | 1.11 (0.97–1.28) | 0.138 |
| rs17828907 | 869 | 892 | 908 | 956 | 1777 | 1848 | ||||||
| AA | 750 (86.3) | 738 (82.7) | 1.00 | 768 (84.6) | 814 (85.2) | 1.00 | 1518 (85.4) | 1552 (84.0) | 1.00 | |||
| AT | 111 (12.8) | 150 (16.8) | 0.71 (0.54–0.93) | 0.027 | 131 (14.4) | 133 (13.9) | 1.03 (0.78–1.37) | 0.773 | 242 (13.6) | 283 (15.3) | 0.86 (0.71–1.05) | 0.144 |
| TT | 8 (0.9) | 4 (0.5) | 2.23 (0.65–7.64) | 0.120 | 9 (1.0) | 9 (0.9) | 0.91 (0.34–2.40) | 0.817 | 17 (1.0) | 13 (0.7) | 1.38 (0.65–2.91) | 0.302 |
| AT+TT | 119 (13.7) | 154 (17.3) | 0.74 (0.57–0.97) | 0.030 | 140 (15.4) | 142 (14.8) | 1.02 (0.78–1.34) | 0.871 | 259 (14.6) | 296 (16.0) | 0.88 (0.73–1.07) | 0.204 |
| Additive model | — | — | 0.81 (0.63–1.05) | 0.106 | — | — | 1.01 (0.79–1.30) | 0.925 | — | — | 0.92 (0.77–1.09) | 0.337 |
Adjusting for age, age at menarche, and menopausal status.
According to the findings from stage I, rs2842346 and rs17828907 were selected into stage II for validation. However, the association between these two SNPs and the risk of breast cancer was not validated in stage II (rs2842346 CT/TT vs. CC: adjusted OR=0.95, 95% CI=0.76–1.20; additive model: adjusted OR=0.99, 95% CI=0.81–1.22; rs17828907 AT/TT vs. AA: adjusted OR=1.02, 95% CI=0.78–1.34; additive model: adjusted OR=1.01, 95% CI=0.79–1.30). Likewise, we also found that neither of these two SNPs was significantly associated with breast cancer risk when two stages were pooled into one (rs2842346 CT/TT vs. CC: adjusted OR=1.09, 95% CI=0.94–1.28; additive model: adjusted OR=1.11, 95% CI=0.97–1.28; rs17828907 AT/TT vs. AA: adjusted OR=0.88, 95% CI=0.73–1.07; additive model: adjusted OR=0.92, 95% CI=0.77–1.09). Additionally, we further performed stratification analyses based on age, age at menarche, age at first live birth, menopausal status, ER and PR status, and did not find any significant difference with regard to these two SNPs in different strata (p>0.10 for heterogeneity tests, Table 4).
Table 4.
Stratified Analysis on the Associations Between Rs2842346 and Rs17828907 and Risk of Breast Cancer in the Combined Dataset
| |
rs2842346 (Case/control) |
rs17828907 (Case/control) |
||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | CC (%) | CT/TT (%) | ORa | pb | AA (%) | AT/TT (%) | ORa | pb |
| Age | ||||||||
| <50 | 723/774 | 241/239 | 1.06 (0.86–1.32) | 0.731 | 842/851 | 133/156 | 0.83 (0.63–1.08) | 0.487 |
| ≥50 | 575/640 | 223/201 | 1.12 (0.89–1.41) | 676/701 | 126/140 | 0.95 (0.73–1.25) | ||
| Menopausal status | ||||||||
| Premenopausal | 610/738 | 224/224 | 1.20 (0.96–1.50) | 0.175 | 733/811 | 110/146 | 0.81 (0.61–1.07) | 0.291 |
| Postmenopausalc | 567/637 | 188/206 | 0.96 (0.76–1.21) | 637/705 | 122/136 | 1.00 (0.76–1.31) | ||
| Age at menarche | ||||||||
| <15 | 737/541 | 269/188 | 1.05 (0.85–1.31) | 0.529 | 862/610 | 157/116 | 0.94 (0.72–1.23) | 0.441 |
| ≥15 | 537/865 | 182/251 | 1.16 (0.93–1.45) | 626/936 | 95/177 | 0.81 (0.62–1.06) | ||
| Age at first live birth | ||||||||
| <25 | 619/906 | 226/306 | 1.04 (0.84–1.28) | 0.169 | 734/1012 | 121/196 | 0.83 (0.65–1.07) | 0.265 |
| ≥25 | 597/473 | 209/118 | 1.32 (1.01–1.72) | 686/498 | 124/90 | 1.04 (0.76–1.41) | ||
| ER status | ||||||||
| ER+ | 574/1414 | 213/440 | 1.09 (0.89–1.33) | 0.903 | 550/1552 | 88/296 | 0.97 (0.76–1.23) | 0.327 |
| ER− | 469/1414 | 164/440 | 1.11 (0.90–1.38) | 674/1552 | 124/296 | 0.81 (0.62–1.06) | ||
| PR status | ||||||||
| PR+ | 582/1414 | 211/440 | 1.08 (0.88–1.31) | 0.716 | 679/1552 | 123/296 | 0.96 (0.75–1.21) | 0.462 |
| PR− | 457/1414 | 168/440 | 1.14 (0.92–1.41) | 543/1552 | 89/296 | 0.84 (0.65–1.10) | ||
Adjusted by age, age at menarche, and menopausal status where appropriate.
P for heterogeneity.
Postmenopausal status for natural menopause.
Discussion
This study used the fine-mapping analysis to assess the relationship between common genetic variants at 14q24.1 and the risk of breast cancer in a Chinese population and found that none of the tagging SNPs was significantly associated with breast cancer risk. The findings suggested that susceptibility loci for breast cancer risk identified by European GWAS were not applicable to other populations such as Chinese.
The 14q24.1 was first identified as a susceptible region of breast cancer in GWAS conducted by Thomas et al. (2009), which had been further confirmed by the two following studies in Europeans (Turnbull et al., 2010; Li et al., 2011). Li et al. (2011) found that the variant allele (A) of rs999737 was significantly associated with a reduced risk of breast cancer by 11% (OR=0.89, 95% CI: 0.81–0.97) after the combined analysis of three GWASs, including a total of 2,702 breast cancer cases and 5,726 controls of European ancestry. Similarly, Turnbull et al. (2010) reported a per-allele OR of 0.89 (95 CI%: 0.83–0.95) for the rs999737-A allele associated with the risk of breast cancer in the UK population with 3,659 cases and 4,897 controls. Rs99937 is located in intron 12 of RAD51L1 (also known as RAD51B) gene encoding the longest transcript isoform 3. RAD51L1, one of the evolutionarily conserved proteins in the RAD51 family, forms a stable heterodimer with another family member RAD51C, which further interacts with the other family members, such as RAD51, XRCC2, and XRCC3 (Miller et al., 2002; Lio et al., 2003). RAD51L1 has been shown to be crucial in the maintenance of genome stability by catalyzing the double-strand break repair though the process of the homologous-recombination pathway (Li and Heyer, 2008). However, it is still unknown which variants in the LD region at the intron 12 of RAD51L1 are causal for breast cancer and how these variants exert their effect on the development of breast cancer. Rs999737 was common in populations of European descent (MAF: 0.242) but very rare (MAF: 0.011) in the Chinese population according to the HapMap database, suggesting that the results from the European population in the reported GWASs (Lichtenstein et al., 2000; Yang et al., 2005) might not be applicable to the Chinese population.
In our study, although we found that two SNPs (rs2842346 and rs17828907) in 14q24.1 were significantly associated with the risk of breast cancer in stage I, neither of them remained significant after the second stage validation and the combined analysis, suggesting that common SNPs in 14q24.1 region might not play a role in breast cancer risk in Chinese women as it did in Europeans. The discrepancy between different studies may be due to a difference in structures of LD across different populations. For example, we found that both the loci (rs2842346 and rs17828907) identified in stage I had a weak LD with rs999737 in the Chinese population (r2=0 and 0.19, respectively); however, rs17828907 were in strong LD with rs999737 in the European population (r2=0.91), which indicated the genetic heterogeneity between Chinese and European populations. Furthermore, a recent GWAS (Wang et al., 2010) with 14,123 BRCA1 and 8,053 BRCA2 mutation carriers of European ancestry reported that rs999737 was not associated with breast cancer risk for either BRCA1 or BRCA2 mutation carriers (p-trend=0.27 and 0.30, respectively), indicating that the effect of SNPs located in 14q24.1 on breast cancer risk could be affected by the status of BRCA1 or BRCA2 mutations. However, no such information was available from three other GWASs (Lichtenstein et al., 2000; Yang et al., 2005) and our study.
There are several limitations that need to be addressed in this study. First, the results of two SNPs (rs2842346 and rs17828907) were not consistent between stage I and stage II, which may be induced by some reasons. On one hand, the significant results in stage I might be false positive and, thus, we could not validate that in stage II; on the other hand, although we used the two-stage design to reduce the false positive, the relative small sample size might reduce the power of the study and miss some true associations. For example, the power values of rs2842346 and rs17828907 in the dominant model were 85% and 93% for one-stage design, and 66% and 77% for the validation stage, respectively. When the two stages were combined into one for joint analysis, the power values of rs2842346 and rs17828907 were 66% and 77%, respectively. Second, this fine-mapping study just focused on the common genetic variants (MAF>0.05) included in the HapMap database. It is possible that other genetic variants, for example, rare variants, may be functional but are not well tagged by the SNPs selected in this study. Third, we did not detect BRCA1 or BRCA2 mutations and evaluate the effect of such mutations on our results. Therefore, further studies with larger samples or detailed information of BRCA1 and BRCA2 mutations are needed to validate our findings.
Conclusions
To our knowledge, this is the first study that focuses on associations between SNPs in the LD region (chr14:68044030–68121653), including rs999737 and breast cancer risk in the non-European population. On the basis of a fine-mapping study in a Chinese population, we found that none of the common SNPs at 14q24.1 was associated with the risk of breast cancer, which would call for replication in other larger studies.
Acknowledgments and Funding
The authors thank the study participants and research staff for their contributions and commitment to this study. This work was supported in part by National Natural Science Foundation of China (grant number 81071715), Key Grant of Natural Science Foundation of Jiangsu Higher Education Institutions (09KJA330001), The Program for Changjiang Scholars, and Innovative Research Team in University (IRT0631).
Authors' Contributions
H.S., Z.H., and H.M. designed the study and prepared the manuscript. J.D performed the experiments with assistance from Z.Q., and J.C., H.M., and G.J. conducted the analysis with assistance from J.D., H.L., and S.W. helped collect and organize the clinical data. H.M., H.S., and X.W. helped finalize the manuscript. All authors read and approved the final manuscript.
Disclosure Statement
The authors declare that they have no competing interests.
References
- Breast Cancer Association Consortium. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst. 2006;98:1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- Han J. Jiang T. Bai H. Gu H. Dong J. Ma H., et al. Genetic variants of 6q25 and breast cancer susceptibility: a two-stage fine mapping study in a Chinese population. Breast Cancer Res Treat. 2011;129:901–907. doi: 10.1007/s10549-011-1527-x. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P. Holm N.V. Verkasalo P.K. Iliadou A. Kaprio J. Koskenvuo M., et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Li X. Heyer W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Humphreys K. Heikkinen T. Aittomäki K. Blomqvist C. Pharoah P.D., et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- Lio Y.C. Mazin A.V. Kowalczykowski S.C. Chen D.J. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- Miller K.A. Yoshikawa D.M. McConnell I.R. Clark R. Schild D. Albala J.S. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J Biol Chem. 2002;277:8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- Ma H. Hu Z. Zhai X. Wang S. Wang X. Qin J., et al. Joint effects of single nucleotide polymorphisms in P53BP1 and p53 on breast cancer risk in a Chinese population. Carcinogenesis. 2006;27:766–771. doi: 10.1093/carcin/bgi295. [DOI] [PubMed] [Google Scholar]
- Manolio T.A. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Thomas G. Jacobs K.B. Kraft P. Yeager M. Wacholder S. Cox D.G., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C. Ahmed S. Morrison J. Pernet D. Renwick A. Maranian M., et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–547. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Pankratz V.S. Fredericksen Z. Tarrell R. Karaus M. McGuffog L., et al. Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2010;19:2886–2897. doi: 10.1093/hmg/ddq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Parkin D.M. Ferlay J. Li L. Chen Y., et al. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev. 2005;14:243–250. [PubMed] [Google Scholar]