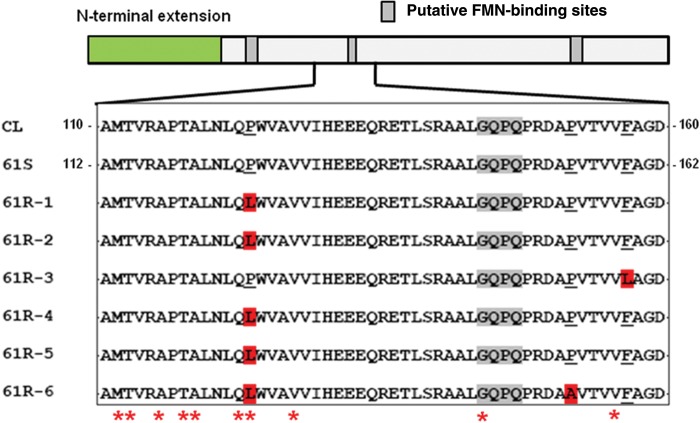
Figure 2.
Mutations in TcNTR from benznidazole-resistant Trypanosoma cruzi. The TcNTR schematic identifies the amino terminal extension (excluded from recombinant proteins) and the location of putative flavin mononucleotide (FMN)–binding regions inferred by analogy with Escherichia coli nfsB [30]. Full-length copies of TcNTR from 61R resistant clones were amplified and sequenced. Differences in the amino acid sequence as compared to the parental TcNTR (61S) were restricted to a single region and are highlighted in red. Several 61S clones were sequenced, but no differences were identified. The sequence in this region of 61S TcNTR (residues 112–162) is identical to that in the genome strain CL Brener (GenBank accession no. XP_810645). The corresponding CL Brener TcNTR residues are numbered 110–160 because of an insertion or deletion in the amino terminal domain. Mutations in the corresponding region of E. coli nfsB that confer nitrofurantoin resistance are indicated by asterisks [31].