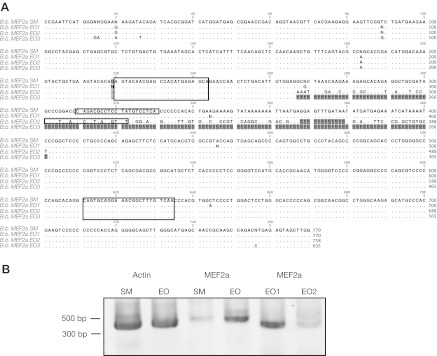
Fig. 6.
Differential expression of MEF2A. The discovery of differentially expressed MEF2A 3′ UTRs in EO prompted further subcloning and sequencing of all MEF2 genes expressed in EO and SM (see Materials and methods). (A) Subcloning revealed multiple sequences of MEF2a expressed in SM (B.b. MEF2a SM1) and EO (B.b. MEF2a EO1–3). These sequences were largely similar except for a region from 264–401 bp that was variable, corresponding to exon 5 of MEF2a in D. rerio. One of the transcripts (B.b. MEF2a EO3) completely lacked this region (gray squares indicate ‘missing’ bases). Using MEF2a-specific primers (large boxed regions), we amplified all MEF2A transcripts expressed in EO to determine which of these sequence variants were expressed in EO and SM. In addition, we developed primers to amplify alternative transcript-specific forms (small boxed regions). (B) RT-PCR comparing actin and MEF2A expression reveals the upregulation of MEF2A in EO. Transcript-specific RT-PCR is consistent with B.b. MEF2a EO1 expressed in high abundance and EO2 expressed in low abundance in EO. Using the above primers, 27 cycles failed to produce an EO3-sized splice variant.