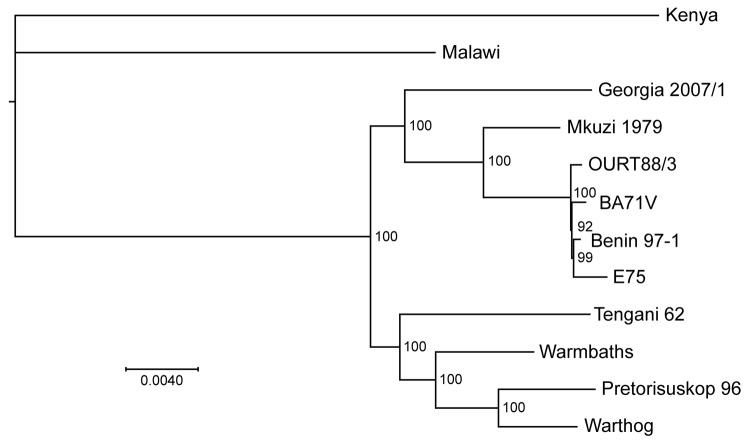
Figure 1.
Comparison of the Georgia 2007/1 African swine fever virus (ASFV) isolate genome with those of other ASFV isolates. ASFV phylogeny midpoint was rooted in a neighbor-joining tree on the basis of 125 conserved open reading frame regions (40,810 aa) from 12 taxa. Node values show percentage bootstrap support (n = 1,000). The isolates shown and accession numbers are Kenya AY261360, Malawi Lil20/1 AY261361, Tengani AY261364, Warmbaths AY261365, Pretorisuskop AY261363, Warthog AY261366, Warmbaths AY261365, Mkuzi AY261362, OurT88/3 a.m.712240, BA71V NC_001659, Benin97/1 a.m.712239, and E75 FN557520. Scale bar indicates nucleotide substitutions per site.