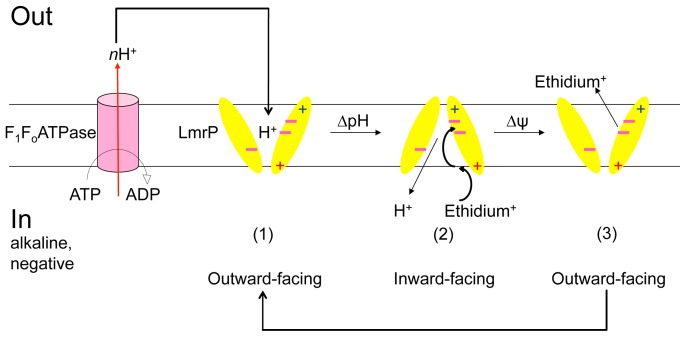
Figure 11. Alternating access model for Δp-dependent ethidium efflux by LmrP.
Proton extrusion by the F1FO-H+ ATPase (in purple) in cells generates a Δp (interior negative and alkaline) across the cytoplasmic membrane. The N- and C-terminal halves of LmrP are represented by yellow ellipses. Minus signs in pink represent the catalytic carboxylates (D142, D235 and E327) that are involved in substrate/proton interactions. During electrogenic efflux via proton-ethidium antiport, carboxylates are protonated in the outward-facing conformation (1), which triggers a ΔpH-dependent conformational switch to the inward-facing conformation (2). Drug binding and proton release at the inside surface will facilitate a second conformational switch in which dissociated carboxylates reorient back to the outward-facing conformation (3) in a Δψ-dependent fashion. Conformation (3) might be the same as conformation (1). K357 (red plus sign, TMH 11) and R260 (blue plus sign, TMH 8) are located on the outer surface of LmrP, near the phospholipid head group region of the inner leaflet (K357) and outer leaflet (R260) of the membrane. In our experiments, alanine replacements at position 357 and 260 shift roughly equal populations of inward-facing and outward-facing LmrP towards outward-facing (K357A) and inward-facing (R260A) populations.