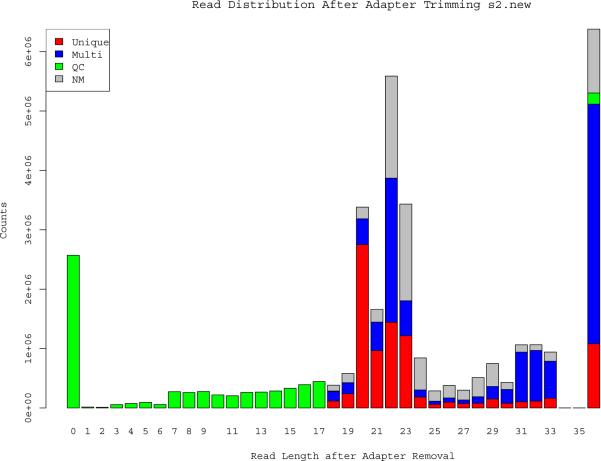
Figure 1.
Size-distribution of the small-RNA reads in one representative sample. The mode read length post adapter trimming occurs at 22bp which is expected from reads generated from miRNAs. The majority of sequences that did not read into the Illumina adapters aligned to tRNAs and rRNAs. Each read was classified as mapping uniquely to the human genome (Unique, red); having multiple alignments to the genome (Multi, blue); failing quality control (QC, green); or not mapping to the genome (NM, grey). The majority of the multiple aligned 20 – 23 bp reads correspond to miRNA families, which were collapsed into a single miRNA species for subsequent analyses.