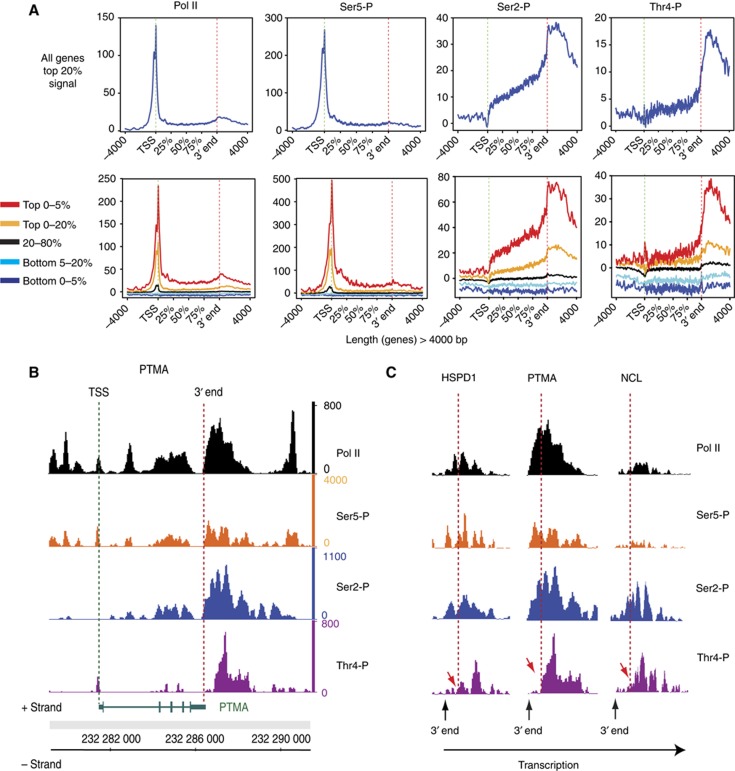
Figure 5.
Genome-wide Pol II and CTD phosphorylation profiles. (A) Diagram summarizing the average transcription unit with representative clusters for Pol II, Ser5-P, Ser2-P, and Thr4-P occupancy profiles across the genome using average transcription unit analysis (see Materials and methods). The top panel represents the profiles for the top 20% of the signal for each Pol II population and the bottom panel various level of binding depending on the signal range. The profiles are built for genes >4 kb in length, which represent the large majority of the protein-coding genes. (B) Protymosin-alpha (PTMA) gene is an example with high Thr4-P levels in the 3′ region of the gene. (C) HSPD1, PTMA, and NCL are examples for Thr4 phosphorylation subsequent to Ser2 phosphorylation at the 3′ end of genes. Normalized ChIPseq signals for each experiment are shown. Genes on positive (+) and negative (−) strand are indicated below the ChIPseq lane. Dashed lines indicate the position in the 3′ region of genes with increase of Thr4-P (red arrows).