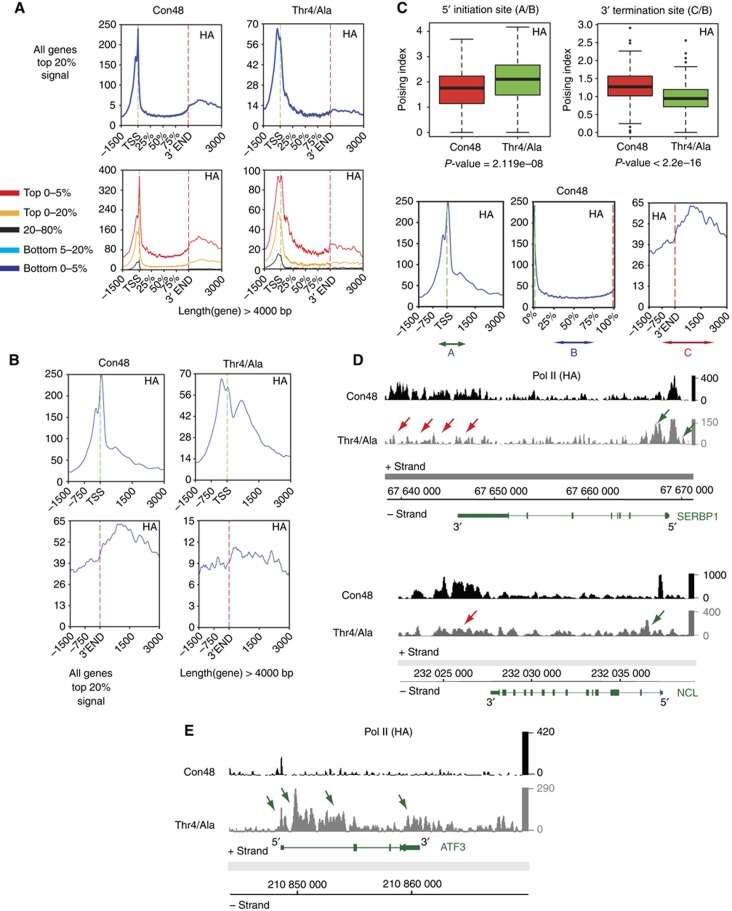
Figure 6.
Genome-wide profiles of CTD mutants Con48 and Thr4/Ala. (A) Average transcription unit occupancy profiles with representative clusters for HA-tagged CTD mutants. Genes with length >4 kb are represented with different selections of signal intensities (different percentages are represented by different colours). (B) Distribution of Pol II upstream and downstream of TSS and 3′ end of genes in Thr4/Ala mutant cell line for the top 20% of binding signal of genes >4 kb. (C) Boxplots of the poising of Pol II at 5′ initiation site and 3′ termination site. In the mutant Thr4/Ala, the poising index is increasing at 5′ whereas it is decreasing at 3′. The profile at bottom shows the areas used for calculating the poising: A=−500/+1000 at TSS, B=50% to end of gene body, C=0–3000 after 3′. Poising at termination site (TS): A/B, poising at 3′: C/B. Genes with Top 5% of binding signal were considered. The significance of the difference of the median was assessed using a non-parametric Mann–Whitney–Wilcoxon test and returned significant P-values of 2.119e−08 and <2.2e−16 for the TSS and 3′ end, respectively. (D) Example of genes reflecting an altered Pol II distribution (SERBP1 and NCL) in the Thr4/Ala mutant. Red arrows indicate reduced Pol II signal at the 3′ end and green arrows increased Pol II at TSSs. More individual examples are shown in Supplementary Figure S7. For all examples shown here and further in the manuscript, DNA is oriented with the chromosomal coordinates direction so that the plus strand shows genes in the 5′–3′ and the minus strand in the 3′–5′ orientation. (E) Example for the minority of genes, ATF3, reflecting an increased Pol II density on the gene body and after the 3′ end, when Thr4 is mutated to alanine.