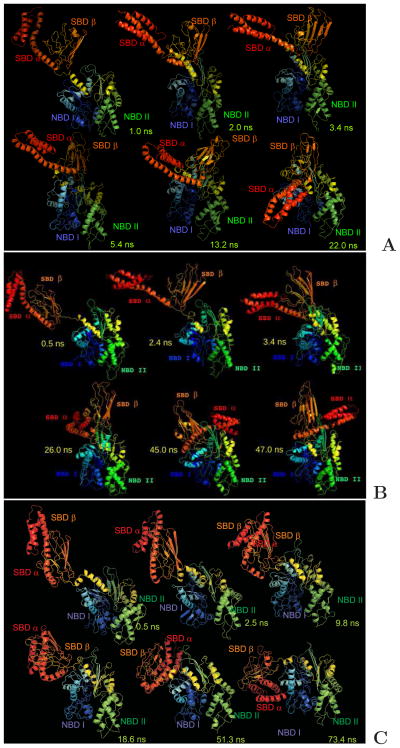
Fig. 6.
Snapshots from representatives of the three types of canonical MD trajectories of 2KHO run with the UNRES force field that are discussed in the text. (A) type I opening-binding trajectory, in which SBD opening is accompanied by binding of SBD-α to NBD-I, resulting in conformations corresponding to that of ATP- and co-chaperone bound structures of yeast Sse1 [PDB IDs: 3C7N8 or 3D2E1],1,9 in which SBD-β binds to NBD from the back side and SBD-α runs from the back to the front of NBD, binding to NBD-I. (B) type II opening-binding trajectory which results in structures with SBD-β bound to the front side of NBD-II and SBD-α bound to the back side of NBD-I. (C) Binding-only trajectory, in which whole SBD binds to NBD without opening. It should be noted that the UNRES simulation time is about 3 orders of magnitude smaller than the actual time.24 All trajectories were obtained in the simulations with the RI-RII type restraints, i.e., without restraining the relative motion of the NBD-I and NBD-II domains. The drawings were done with PyMOL (http://www.pymol.org) and the backbones of the UNRES conformations were converted to all-atom representation by using the knowledge-based algorithm developed by Gront et al.43 to enable to draw the structures in ribbon representation.