Abstract
Twin imaging studies have been valuable for understanding the relative contribution of the environment and genes on brain structures and their functions. Conventional analyses of twin imaging data include three sequential steps: spatially smoothing imaging data, independently fitting a structural equation model at each voxel, and finally correcting for multiple comparisons. However, conventional analyses are limited due to the same amount of smoothing throughout the whole image, the arbitrary choice of smoothing extent, and the decreased power in detecting environmental and genetic effects introduced by smoothing raw images. The goal of this article is to develop a two-stage multiscale adaptive regression method (TwinMARM) for spatial and adaptive analysis of twin neuroimaging and behavioral data. The first stage is to establish the relationship between twin imaging data and a set of covariates of interest, such as age and gender. The second stage is to disentangle the environmental and genetic influences on brain structures and their functions. In each stage, TwinMARM employs hierarchically nested spheres with increasing radii at each location and then captures spatial dependence among imaging observations via consecutively connected spheres across all voxels. Simulation studies show that our TwinMARM significantly outperforms conventional analyses of twin imaging data. Finally, we use our method to detect statistically significant effects of genetic and environmental variations on white matter structures in a neonatal twin study.
Keywords: Heritability, Multiscale adaptive regression model, Smooth, Structural equation model, Twin study
1 Introduction
The typical design of a twin study compares the similarity of monozygotic twins (MZ), who share the same genetic polymorphisms, to that of dizygotic twins (DZ), who share an average of 50% of their genetic polymorphisms. Under the equal environment assumption across zygosities, the known differences in genetic similarity allows us to dissect the effects of genes and environment on a known phenotype, such as total brain volume and volumes of subcortical structures. Thus, twin neuroimaging studies have been a valuable source of information for evaluating the inheritance of brain structure and function by disentangling genetic factors from environment [1, 2, 3, 4, 5, 6, 7, 8]. For instance, high heritability was found in different brain volumes including intracranial volume, total brain volume, and global gray and white matter volumes [9, 3, 10]. Moreover, cortical thickness in the sensorimotor cortex, middle frontal cortex, and anterior temporal cortex were found to be influenced by genetic factors [5, 11].
The voxel-wise methods for analyzing twin neuroimaging data on a two-dimensional (2D) surface (or in a three-dimensional (3D) volume) are sequentially executed in three steps. The first step is to use standard smoothing methods to spatially smooth the imaging data [5, 6, 7]. These smoothing methods are commonly independent of the imaging data and apply the same amount of smoothing throughout the whole image. See, for example, [12] for overviews of some smoothing methods in the neuroimaging literature. The second step involves fitting statistical models to imaging measures from all twin pairs at each voxel or surface location separately to generate a parametric map of test statistics (or p–values) [13, 14]. The most commonly used models are structural equation models, which primarily decompose the observed phenotypic variance into additive genetic, dominant genetic, common environmental components, and random noise [15, 16]. The third step is to compute adjusted p-values in order to correct for testing multiple hypotheses across thousands to millions of locations using multiple comparison correction methods (e.g., random field theory (RFT), false discovery rate (FDR), or permutation methods)[17, 18, 19, 20, 21, 22].
The existing voxel-wise methods based on Gaussian kernel smoothing and structural equation models have two major limitations for analyzing twin neuroimaging data, which underscore the great need for further methodological development. It is well known that the commonly used Gaussian kernel for smoothing imaging data usually blurs the image data near the edges of the significant regions, which can dramatically increase the numbers of false positives and negatives. As shown in [23, 24], the existing voxel-wise methods can suffer from the arbitrary choice of smoothing extent in the initial smoothing step, which leads to the large number of false positive and false negative results. An overview of different smoothing methods that are used in the neuroimaging literature can be found in [12]. Generally, most methods for smoothing raw twin imaging data take a weighted average of the imaging data at voxels in the neighborhood of each voxel, thus are primarily associated with the mean structure of imaging data. Such a way of smoothing raw twin imaging data can change the variance structure of imaging data, which is primarily associated with genetic and environmental factors [25, 16, 15, 13, 14]. Thus, directly smoothing twin imaging data can introduce substantial bias in estimating these factors and lead to a dramatic increase of the numbers of false positives and negatives. We will investigate the consequence of smoothing twin imaging data by theoretical arguments in the Appendix and using simulated data in Section 3. Recently, [26] examined how the overall significance of voxel-wise effects varied with respect to full width at half maximum(FWHM) of the Gaussian smoothing applied to the fractional anisotropy (FA) images. Their results have shown that raw FAmaps with no smoothing yielded the greatest sensitivity to detect gene effects. Our results give a theoretical justification for their results. In general, directly smoothing raw twin imaging data by using Gaussian kernel should be avoided in practice.
As pointed out by [20] and many others, these voxel-wise methods essentially treat all voxels as independent units in the modeling stage, and they explicitly model the spatial coherence and incorporate the information that spatially contiguous regions of effect with rather sharp edges may exist in neuroimaging data. At least two groups of statistical methods have been developed to characterize spatial dependence in neuroimaging data. The first group of methods is to explicitly incorporate the spatial non-independence in the p-value or testing statistic images. These methods usually combine strength from both the magnitude of testing statistic and cluster extent, where the cluster is defined as the number of contiguous signiificant voxels above a specific threshold [20, 21, 22].
Instead of solely utilizing information in the p-value or testing statistic images, the other group of methods explicitly incorporates the spatial dependence in their models. It is common to use conditional autoregressive (CAR), Markov random field (MRF), and other spatial correlation priors to characterize spatial dependence among spatially connected voxels [27, 28, 29, 30, 31, 32]. In addition, a wealth of image denoising and restoration methods has been developed for pre-processing a single image in the literature. An overview of image denoising and restoration methods has been given in [33, 34] and references therein. For instance, Polzehl et al.[35, 36] developed a propagation separation(PS) approach to adaptively and spatially smooth images from a single image without detecting edge curves/surfaces explicitly. In Qiu et al.[37, 38], novel three-dimensional (3D) denoising procedures were proposed to nonparametrically estimate a 3-D jump surface from noisy data. In Tabelow et al. [39, 40, 41], the original PS idea was adapted to develop a multiscale adaptive linear model to adaptively and spatially denoise functional magnetic resonance imaging (fMRI) and diffusion tensor images (DTI) from a single subject. Under the linear model for fMRI, In Polzehl et al.[42], a structural adaptive segmentation algorithm was developed to integrate the signal detection with noise detection in one procedure. In Li et al.[43], a multiscale adaptive regression model(MARM) was proposed to integrate the PS approach [35, 36] with general statistical modeling at each voxel for spatial and adaptive analysis of neuroimaging data from multiple subjects in various study designs.
The aim of this article is to develop a pipeline, called TwinMARM, for the spatial and adaptive analysis of twin neuroimaging data. TwinMARM integrates adaptive estimation and testing in a single framework. TwinMARM consists of two stages of multiscale adaptive regression models (MARM). Each stage of TwinMARM constructs hierarchical nested neighborhoods with increasing radii at all voxels, adaptively generates weighted quasi-likelihood functions, and efficiently utilizes available information to estimate parameters. Particularly, in the first stage, weighted quasi-likelihood functions are constructed to capture the mean structure of imaging data in order to establish the relationship between twin imaging data and a set of covariates of interest, such as age and gender. Subsequently, in the second stage, a set of new weighted quasi-likelihood functions is built that utilizes the variance structure of imaging data to disentangle relative contributions of environment and genes on brain structures and their functions.
Compared to all existing methods including MARM and PS, we make several major contributions. TwinMARM represents a novel generalization of MARM and PS. Specifically, PS has been primarily used to solely average imaging intensity, which improves the accuracy in estimating the mean structure of imaging data [42, 39, 40, 41], but we are more interested in variance structure of imaging data in Twin studies. One may directly apply MARM to the structural equation model (SEM) described in Section 2, but we have found that it is computationally difficult to directly optimize the weight likelihood function for SEM. Moreover, the existing MARM and PS methods can only smooth all unknown parameters (or a subset of parameters) simultaneously, since the weights used in MARM and PS depend the weighted distance of the whole parameter vectors. However, we cannot estimate the mean and variance structure of twin imaging data simultaneously. At the same time, extending MARM and PS to smoothing individual parameters is an unsolved problem, since it is very difficult to approximate the covariance matrix of the smoothed parameter estimates obtained from MARM and PS. Fortunately, due to the special model structure for twin data, we are able to find a novel approach, which allows us to apply MARM twice to independently smooth the mean and variance structure of twin imaging data. Theoretically, we have shown in the Appendix that our approach is valid asymptotically. TwinMARM slightly increases the amount of computational time in computing parameter estimates and test statistics, whereas it substantially outperforms the voxel-wise approach in increasing the accuracy of parameter estimates and the power of test statistics. Practically, TwinMARM developed here is applicable to different imaging modalities including DTI, fMRI, and MRI among many others. Particularly, TwinMARM gives a solution to the issue raised in [26]. Due to the importance and popularity of twin imaging studies, TwinMARM will have wide and important applications in the analysis of imaging, genetic, and clinical data collected from twin studies.
2 Methods
2.1 Structural Equation Model
Suppose we observe imaging measures and clinical variables from n1 MZ twin pairs and n2 = n − n1 DZ twin pairs. Imaging measures can be volumes of anatomical regions, the shape representation of the surfaces of cortical or subcortical structures, fMRI signals, diffusion tensors, and tensor-derived measures [44, 45, 46, 47, 48, 49, 50, 5, 6, 7, 8]. Clinical variables may include demographic and environmental variables. Specifically, for the j-th subject in the i-th twin pair, we observe an NV × 1 vector of imaging measures, denoted by Yij = {yij(v) : v ∈
 }, and a k × 1 vector of clinical variables xij = (xij1, ···, xijk)T for i = 1, ·, n and j = 1, 2, where xij1 is commonly set as 1 and
}, and a k × 1 vector of clinical variables xij = (xij1, ···, xijk)T for i = 1, ·, n and j = 1, 2, where xij1 is commonly set as 1 and
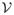 and v, respectively, represent a specific brain region and a voxel in
and v, respectively, represent a specific brain region and a voxel in
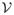 . For notational simplicity, we only consider univariate measure.
. For notational simplicity, we only consider univariate measure.
At a specific voxel v, we consider the structural equation model
| (1) |
for j = 1, 2 and i = 1, ···, n = n1 + n2, where β(v) = (β1(v), ···, βk(v))T is a k × 1 vector representing unknown regression coefficients, aij(v), dij(v), ci(v) and eij(v) are, respectively, the additive genetic, dominance genetic, common environmental, and residual effects on the i-th twin pair. It is common to assume that aij(v), dij(v), ci(v) and eij(v) are independently normally distributed with means 0 and variances , and , respectively [16, 15, 13, 14, 51]. Moreover, Cov(ai1(v), ai2(v)) equals for MZ twins and for DZ twins, while Cov(di1(v), di2(v)) equals for MZ twins and for DZ twins [52]. See a path diagram of model (1) in Figure 1. Due to an identifiability issue, we may not be able to dissect the additive genetic effect, the dominance genetic effect, and the common environmental effect from the residual environmental effect [16, 15, 13, 14]. It is common to consider two simpler models. One is the ACE model, which only includes additive genetic, common environmental, and residual effects. The other is the ADE model, which includes additive and dominance genetic, and residual effects.
Fig. 1.
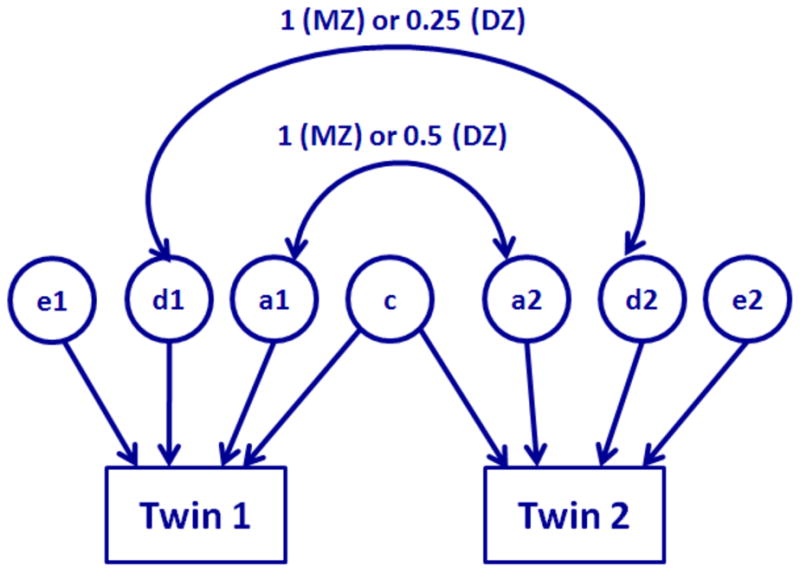
Diagram for the structural equation model for twin data. The correlation of additive effects (a1, a2) is 1 for MZ twin and 0.5 for DZ twin. The correlation of dominant effects (d1, d2) is 1 for MZ twin and 0.25 for DZ twin. The twin share the same common environmental effect (c). Residual effects (e1, e2) for twin are not correlated.
Without loss of generality, we focus on the ACE model and drop the assumption of normality for aij(v), dij(v), ci(v) and eij(v) from now on, since TwinMARM is independent of such assumption. For the ACE model, if we are able to estimate all variances , we can disentangle relative contribution of environment and genetic polymorphisms. Specifically, the genetic heritability at voxel v, denoted by g(v), is defined as the ratio of the genetic variance to the total variance of yij(v) given by
2.2 TwinMARM
We propose the two-stage multiscale adaptive regression method (TwinMARM) for the analysis of twin imaging data and behavioral data as follows. The first stage is to estimate β = {β(v) : v ∈
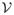 }, while the second stage is to estimate η = {η(v) : v ∈
}, while the second stage is to estimate η = {η(v) : v ∈
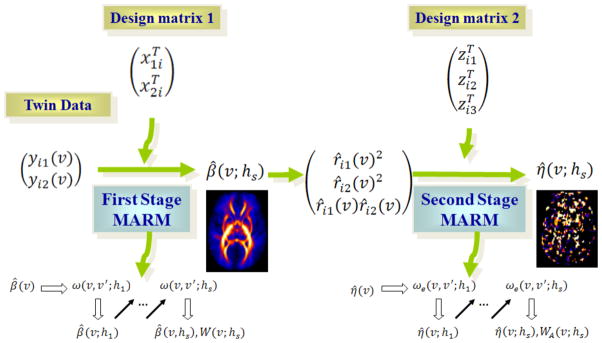
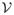 }. In each stage, we reformulate the problem of estimating β (or η) as a regression model and then apply the multiscale adaptive regression model (MARM) in [43], which can be regarded as a generalization of the propogation-seperation (PS) procedure in multiple subjects [35, 53, 36, 39, 40, 41]. The key ideas of MARM and PS are to construct hierarchical nested neighborhoods with increasing radii at all voxels, to adaptively construct weighted quasi-likelihood functions to estimate parameter estimates, and to increase the power of test statistics in detecting subtle changes of brain structures and their functions. A path diagram of TwinMARM is presented in Figure 2.
}. In each stage, we reformulate the problem of estimating β (or η) as a regression model and then apply the multiscale adaptive regression model (MARM) in [43], which can be regarded as a generalization of the propogation-seperation (PS) procedure in multiple subjects [35, 53, 36, 39, 40, 41]. The key ideas of MARM and PS are to construct hierarchical nested neighborhoods with increasing radii at all voxels, to adaptively construct weighted quasi-likelihood functions to estimate parameter estimates, and to increase the power of test statistics in detecting subtle changes of brain structures and their functions. A path diagram of TwinMARM is presented in Figure 2.
Fig. 2.
Diagram for the TwinMARM method for twin data. The first stage is to estimate β = {β(v) : v ∈
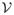 }, while the second stage is to estimate η = {η(v) : v ∈ V}. In each stage, we reformulate the problem of estimating β (or η) as a regression model and then apply the multiscale adaptive regression method (MARM) to spatially and adaptively calculate β̂(v;hs) and η̂(v; hs) and their associated test statistics. Moreover, yij denote imaging measures for twin pairs, xij is a vector of clinical variables, β is the vector of unknown regression parameters, rij are residuals obtained from first stage, η include the variances of additive genetic, common environmental and residual effects, and zij is the design matrix for the second stage. ω(v, v′; hs) is the weight of the first stage, whereas ωe(v, v′; hs) is the weight of the second stage.
}, while the second stage is to estimate η = {η(v) : v ∈ V}. In each stage, we reformulate the problem of estimating β (or η) as a regression model and then apply the multiscale adaptive regression method (MARM) to spatially and adaptively calculate β̂(v;hs) and η̂(v; hs) and their associated test statistics. Moreover, yij denote imaging measures for twin pairs, xij is a vector of clinical variables, β is the vector of unknown regression parameters, rij are residuals obtained from first stage, η include the variances of additive genetic, common environmental and residual effects, and zij is the design matrix for the second stage. ω(v, v′; hs) is the weight of the first stage, whereas ωe(v, v′; hs) is the weight of the second stage.
2.2.1 TwinMARM: Stage I
The first stage is to estimate β(v) and carry out statistical inference on β(v). Specifically, we consider a bivariate regression model given by
| (2) |
where and fi(v) = (fi1(v), fi2(v))T, in which fij(v) = aij(v) + ci(v) + eij(v) for the ACE model. Although fi(v) has different covariance structures for MZ and DZ, respectively, we assume that fi(v) has mean zero and covariance Σf (v) for all i to avoid estimating variance components ( ) in Stage I, which leads a simple procedure for estimating β. We will appropriately account for the misspecification of covariance of fi(v) in our statistical inference on β(v).
Since all components of β(v) are the parameters of interest and Σf (v) can be regarded as nuisance parameters, we first estimate Σf (v) across all voxels and then fix them at their estimated values. Specifically, let ℓ(Yi(v)|xi, β(v), Σf (v)) be the pseudo-likelihood of Yi(v) given by
| (3) |
We calculate the maximum pseudo-likelihood estimate of (β(v), Σf (v)) given by
| (4) |
Let a⊗2 = aaT for any vector a and (β̂(v)(t), Σ̂f (v)(t)) be the estimate of (β(v), Σf (v)) at the t-th iteration. Computationally, at each voxel v, by setting Σ̂f (v)(0) = I2, a 2 × 2 identity matrix, we iteratively update
| (5) |
until that the difference between (β̂(v)(t+1), Σ̂f (v)(t+1)) and (β̂(v)(t), Σ̂f (v)(t)) is smaller than a prefixed constant, say 10−6. From now on, Σf (v) will be fixed at Σ̂f (v) across all voxels v. Statistically, it can be shown that such a substitution has a negligible effect on statistical inference of β.
To estimate β(v) at voxel v, we construct a weighted quasi-likelihood function by utilizing all imaging data in a sphere with radius h at voxel v, denoted by B(v, h). Let ω(v, v′; h) ∈ [0, 1] be a weight function of two voxels and a radius h, which characterizes the similarity between the data in voxels v and v′ such that ∑v′∈B(v,h) ω(v, v′; h) = 1 for all h > 0. If ω(v, v′; h) ≫ 0, it represents that the data in voxels v and v′ are similar to each other, whereas ω(v, v′; h) ≈ 0 indicates that the data in voxel v′ do not contain too much information on β(v). The adaptive weights ω(v, v′; h) play an important role in preventing oversmoothing the estimates of β(v) as well as preserving the edges of significant regions. We utilize all the data {Yi(v′) : v′ ∈ B(v, h)} to construct the weighted log-likelihood function at voxel v at scale h, which is given by
| (6) |
By directly maximizing ℓobs(β(v); h), we can obtain the maximum pseudo-likelihood estimate of β(v), denoted by β̂(v; h), which is given by
| (7) |
where . The covariance matrix of β̂(v; h) can be approximated by
| (8) |
where . Although [39] have obtained the same β̂(v; h) as in expression (7), the covariance estimate of β̂(v; h) in expression (8) has a simple form compared to the corresponding one in [39] with an additional spatial correlation assumption. We have found that such spatial correlation assumption is unnecessary since Σn,2(v; h) has implicitly incorporated the spatial correlation in the data.
Based on β̂(v; h), we can further construct test statistics to examine scientific questions about β(v). For instance, such questions may compare brain structure across different groups (MZ versus DZ) or detect change in brain structure across time [3, 4, 5, 6, 7, 8]. These questions can be formulated as the linear hypotheses about β(v)
| (9) |
where μ = R1β(v), R1 is a r × k matrix of full row rank and b0 is a r × 1 specified vector. We test the null hypothesis H0,μ : R1β(v) = b0 using the Wald test statistic
| (10) |
Under H0,μ, Wμ(v; h) is asymptotically distributed as χ2(r) [43].
2.2.2 Propagation-Seperation procedure
Following the propagation-seperation (PS) procedure proposed in [35, 53, 36], [39, 40, 41] and [43], we adaptively determine {ω(v, v′; h) : v, v′ ∈
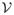 } and then calculate β̂(v; h) and Wμ(v; h) as h increases from 0 to a preset value r0. Since PS and the choice of its associated parameters have been described in details in [35, 53, 36] and [43], we briefly disscuss them here for the sake of simplicity.
} and then calculate β̂(v; h) and Wμ(v; h) as h increases from 0 to a preset value r0. Since PS and the choice of its associated parameters have been described in details in [35, 53, 36] and [43], we briefly disscuss them here for the sake of simplicity.
The key idea of PS is to build a sequence of nested spheres with increasing radii h0 = 0 < h1 < ··· < hS = r0 at each voxel v ∈
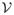 . When h = h1, we extract features from the imaging data as well as {β̂(v) : v ∈
. When h = h1, we extract features from the imaging data as well as {β̂(v) : v ∈
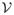 } and compute weights ω(v, v′; h1) at scale h1 for all v, v′ ∈
} and compute weights ω(v, v′; h1) at scale h1 for all v, v′ ∈
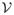 . Then, we adaptively determine ω(v, v′; hs) and update β̂(v; hs) from h1 to hS = r0. A path diagram of the multiscale strategy is given below:
. Then, we adaptively determine ω(v, v′; hs) and update β̂(v; hs) from h1 to hS = r0. A path diagram of the multiscale strategy is given below:
The four key steps of PS including weights adaptation, estimation, stop checking, and inference are presented as follows.
-
In the weights adaptation step (i), we prefix a series{ } of radii with ch ∈ (1, 2), say ch = 1.15. We then set s = 1 and h1 = ch. The adaptive weights are given by
(11) where Kloc(u) = (1−u)+ and Kst(u) = min(1, 2(1−u2))+ according to our experience and previous literature [39, 43], and || · ||2 denotes the Euclidean norm of a vector. Moreover, Dβ(v, v′; hs−1) is set as(12) Following Li et al (2010), we choose Cn = n1/3χ2(k)0.95 for Dβ(v, v′; hs−1) defined in (12), where χ2(k)0.95 is the upper 0.05-percentile of the χ2(k) distribution. The adaptive weight Kst(Dβ(v, v′; hs−1)/Cn) downweights the role of a voxel v′ ∈ B(v; hs) in ℓobs(β(v); hs) if the value of Dβ(v, v′; hs−1) is large. The weight Kloc(||v − v′||2/hs) gives less weight to a voxel v′ ∈ B(v; hs) whose location is far from the voxel v. Since the theoretical justification for choosing various parameters in ω(v, v′; hs) has been discussed in [35, 53, 36] and [43], we omit them here for the sake of simplicity.
In the estimation step (ii), for the radius hs, we calculate β̂(v; hs) given in (7) at each voxel v ∈
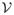 .
.-
In the stop checking step (iii), after the S0–th iteration, we calculate a stopping criterion based on a normalized distance between β̂(v; hS0) and β̂(v; hs) given by
(13) Then, we compare D(β̂(v; hS0), β̂(v; hs)) with a benchmark, denoted by C̃(s), for s > S0. If D(β̂(v; hS0), β̂(v; hs)) is greater than C̃(s), then we set β̂(v, hS) = β̂(v; hs−1), and s = S. If s = S, we go to the inference step (iv). If s ≤ S0 or D(β̂(v; hS0), β̂(v; hs)) ≤ C̃(s) for S − 1 ≥ s > S0, then we set hs+1 = chhs, increase s by 1 and continue with the weights adaptation step (i). Throughout the paper, we set S0 = 3 and C̃(s) = χ2(p)0.8/(s−2)0.9.
In the inference step (iv), when s = S, we compute Wμ(v; hS) for all v ∈
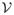 and then apply either FDR or RFT correction to obtain the corrected p-values of Wμ(v; hS) in order to detect significant voxels [19, 20, 21, 22]. Otherwise, we set hs+1 = chhs and continue with the weights adaptation step. The maximal step S can be taken to be relatively small, say 10, and thus the largest spherical neighborhood of each voxel only contains a relatively small number of voxels compared to the whole volume. Throughout the paper, we have used the false discovery rate method under the positive dependency in [17], since the test statistics obtained from MARM satisfy such dependency condition.
and then apply either FDR or RFT correction to obtain the corrected p-values of Wμ(v; hS) in order to detect significant voxels [19, 20, 21, 22]. Otherwise, we set hs+1 = chhs and continue with the weights adaptation step. The maximal step S can be taken to be relatively small, say 10, and thus the largest spherical neighborhood of each voxel only contains a relatively small number of voxels compared to the whole volume. Throughout the paper, we have used the false discovery rate method under the positive dependency in [17], since the test statistics obtained from MARM satisfy such dependency condition.
2.2.3 TwinMARM: Stage II
The second stage is to estimate
and carry out statistical inference on η(v). Given {β̂(v; hS) : v ∈
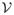 } obtained from Stage I, we compute residuals
for all i, j across all v ∈
} obtained from Stage I, we compute residuals
for all i, j across all v ∈
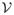 . Then, we consider a trivariate regression model given by
. Then, we consider a trivariate regression model given by
| (14) |
where εi(v) = (εi1(v), εi2(v), εi3(v))T and , in which zi1 = zi2 = (1, 1, 1)T and zi3 equals (0.5, 1, 0)T for DZ and (1, 1, 0)T for MZ. We have given the detailed derivation of model (14) in the Appendix. Moreover, εi(v) is assumed to have mean zero and covariance Σε(v) for all i. Although the homogeneous variance of εi(v) may not be realistic, we can appropriately account for such a misspecified assumption as we did for Σf (v) in Stage I. Since models (2) and (14) are similar in nature, most developments for model (2) can be directly applied to model (14) and then we omit them for the sake of simplicity.
There are several minor differences between models (2) and (14). In model (14), all regression coefficients in η(v) are constrained to be non-negative. Without loss of generality, Σε(v) is assumed to be known since it can be replaced by a consistent estimate. The quasi-likelihood function for η(v) is given by
| (15) |
Let be an unconstrained estimate of η(v) in model (15). By discarding these terms independent of η(v), we can show that ℓobs(η(v); h0) can be simplified into
The constrained estimate of η(v), denoted by η̂(v), is given by
| (16) |
Since the set [0, ∞)3 = [0, ∞) × [0, ∞) × [0, ∞) is a convex cone, η̂(v) is unique [54]. To compute η̂(v), we employ a hinge algorithm for cone projection [54].
Furthermore, similar to (6), we can utilize weights ωe(v, v′; h) to construct a weighted quasi-likelihood function as follows:
| (17) |
Similarly, η̂(v; h) is given by
| (18) |
where η̃(v; h) equals
| (19) |
The same hinge algorithm [54] is used to calculate η̂(v; h).
The second focus common in twin study research is the testing of genetic and environmental influences on brain structures and their functions. See for example, [2], [55, 11], [5], [6], [7], and [8], among many others. Such questions of interest can be formulated as the linear hypotheses about η(v) as follows:
| (20) |
where R2 is an m × 3 matrix. For instance, in an ACE model, R2 equals (1, 0, 0) for genetic effect and (0, 1, 0) for environmental effect. Similar to testing H0,μ, we test H0,A using the Wald test statistic
| (21) |
where Σn(η̂(v; h)) is the covariance matrix of η̂(v; h). The statistic WA(v; h) is asymptotically distributed as a mixture of χ2(1) and χ2(0) distributions under the null hypothesis H0,A, where χ2(0) = 0. However, to test the joint genetic and environmental effects, R2 takes the form of [(1, 0, 0)T(0, 1, 0)T] and WA(v; h) is asymptotically distributed as a mixture of χ2(2), χ2(1) and χ2(0) distributions with the weights depending on the ratio of MZ over DZ [51, 56].
3 Simulation Studies
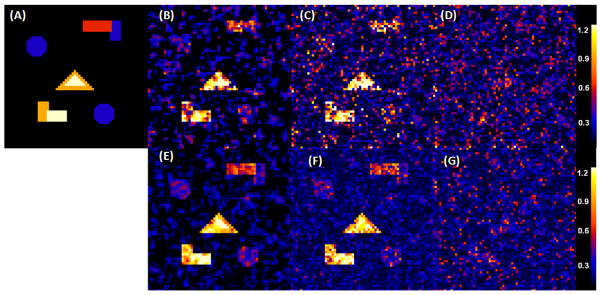
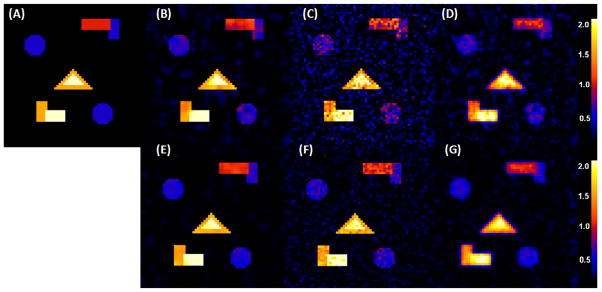
We carried out some simulation studies to examine the finite-sample performance of Twin-MARM. We first simulated MRI measures from n pairs of siblings according to the ACE model, in which β(v) = (β1(v), β2(v), β3(v))T and xij = (x1ij, x2ij, x3ij)T for j = 1, 2. Each family contains only two siblings. Among the n pairs of twins, 60% are identical twins. We set x1ij ≡ 1, generated x2ij independently from a Bernoulli distribution with probability of success 0.5 and generated x3ij independently from the Gaussian distribution with zero mean and unit variance. The x2ij and x3ij were chosen to represent gender identity and scaled age, respectively. We set across all voxels v. For ( ), we divided the 64×64 phantom image into five different regions of interest (ROIs) with different shapes and then varied ( ) as (0, 0), (0.3, 0.5), (0.6, 1), (0.9, 1.5) and (1.2, 2.0) across these five ROIs. The true ( ) was displayed for all ROIs with black, blue, red, yellow, and white colors representing ( and (1.2, 2.0) (Fig. 3(A)). We independently generated ei(v) = (ei1(v), ei2(v))T, ai(v) = (ai1(v), ai2(v))T, and ci(v) from multivariate Gaussian distributions with zero means and covariance matrices as specified in Fig. 1. We set n = 100 and 400.
Fig. 3.
Results from a simulation study of comparing EM and TwinMARM at 2 different sample sizes (n = 100, 400). The first row contains the results for as n = 100: Panel (A) is the ground truth image of , in which five ROIs with black, blue, red, yellow, and white color represent , respectively. Panel (B) is a selected slice of obtained from a simulated dataset by using the TwinMARM method. Panel (C) is a selected slice of obtained from the same dataset as panel (B) by using the voxel-wise EM method. Panel (D) is a selected slice of by using EM, after smoothing the same simulated data set as panel (B). The second row contains panels (E), (F) and (G) as n = 400, which are the corresponding results of panels of panels (B), (C) and (D), respectively.
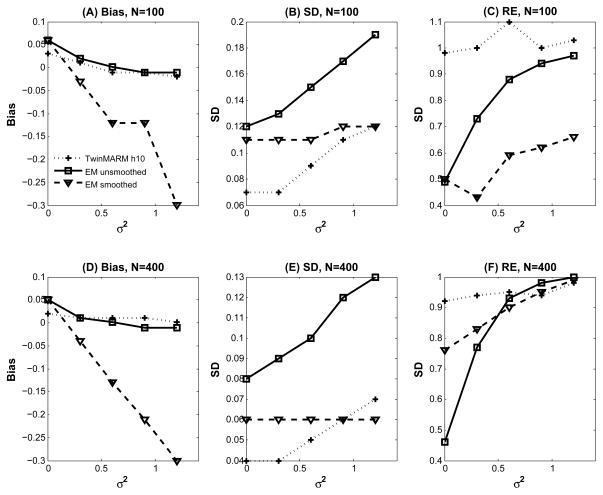
To carry out the analysis, we first fit the structural equation model , in which the correlation pattern of aij(v), ci(v) and eij(v) was specified according to Fig. 1. We calculated the maximum likelihood estimates (MLEs) of at each pixel by using the expectation-maximization (EM) algorithm [57]. Then, we applied the TwinMARM procedure described in Section 2.2 to calculate adaptive parameter estimates across all pixels at 11 different scales. We also smoothed the subjects’ images by using an isotropic Gaussian kernel with FWHM 3 pixels and then applied the EM algorithm to the smoothed data. Furthermore, for (β1(v), ), we calculated the bias, the empirical standard error RMS, the mean of the standard error estimates SD, and the ratio of RMS over SD, abbreviated as RE, at each pixel of all five ROIs based on the results obtained from the 1000 n = 100 and 400 simulated data sets.
For simplicity, we present some selected results for β̂1(v; h) and and their corresponding MLEs obtained from the EM algorithm with smoothed and unsmoothed data. We first considered . The biases of σa(v; h)2 are almost the same for TwinMARM at h10 and EM with unsmoothed data (Fig. 3(B), (C), (E) and (F)), while the bias from EM with smoothed data is greatly increased (Fig. 5(A) and (C) and Fig. 3(D) and (G))). Inspecting the results from the EM algorithm reveals that SD is consistently larger than that obtained from TwinMARM at h10 (Fig. 5(B) and (D)). Overall, for , TwinMARM outperforms EM in terms of smaller RMS (Fig. 3(B), (C), (E) and (F)).
Fig. 5.
Results from a simulation study of comparing EM and TwinMARM at 2 different sample sizes (n = 100, 400). The first row contains the results for as n = 100: Panel (A) is the bias curve of , respectively. Panel (B) is the SD curve of obtained from a simulated dataset by using TwinMARM, EM, and EM with smoothed data. Panel (C) is the ratio of RMS over SD. The second row contains panels (D), (E), and (F) as n = 400, which are the corresponding results of panels of panels (A), (B), and (C) respectively.
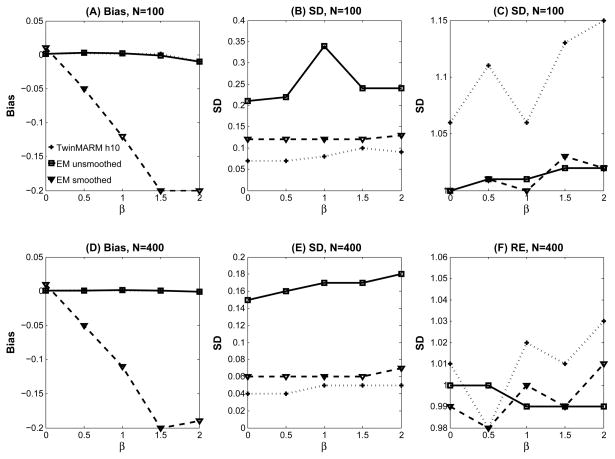
Secondly, we examine β̂1(v; h). The biases of β̂1(v; h) are almost the same for TwinMARM at h10 and EM with unsmoothed data (Fig. 6(A) and (C) and Fig. 4(B), (C), (E) and (F)), while the bias from EM with smoothed data is greatly increased (Fig. 6(A) and (C) and Fig. 4(D) and (G)). Inspecting the results from the EM algorithm reveals that SD is consistently larger than that obtained from TwinMARM at h10 (Fig. 6(B) and (D)). For β̂1(v; h), TwinMARM outperforms EM in terms of smaller RMS (Fig. 4(B), (C), (E) and (F)).
Fig. 6.
Results from a simulation study of comparing EM and TwinMARM at 2 different sample sizes (n = 100, 400). The first row contains the results for β1(v) as n = 100: Panel (A) is the bias curve of β1(v)=0, 0.5, 1.0, 1.5, and 2.0, respectively. Panel (B) is the SD curve of β̂(v) obtained from a simulated dataset by using TwinMARM, EM, and EM with smoothed data. Panel (C) is the ratio of RMS over SD. The second row contains panels (D), (E), and (F) as n = 400, which are the corresponding results of panels of panels (A), (B), and (C) respectively.
Fig. 4.
Results from a simulation study of comparing EM and TwinMARM at 2 different sample sizes (n = 100, 400). The first row contains the results for β1(v), when sample size n = 100: Panel (A) is the ground truth image of β1(v), in which five ROIs with black, blue, red, yellow, and white color represent β1(v)=0, 0.5, 1.0, 1.5, and 2.0, respectively. Panel (B) is a selected slice of β̂1(v; h10) obtained from a simulated dataset by using TwinMARM. Panel (C) is a selected slice of β̂1(v) obtained from the same simulated dataset as panel (B) by using EM. Panel (D) is a selected slice of β̂1(v) obtained by using EM, after smoothing the same simulated data set as panel (B). The second row contains panels (E), (F) and (G) as n = 400, which are the corresponding results of panels of panels (B), (C) and (D), respectively.
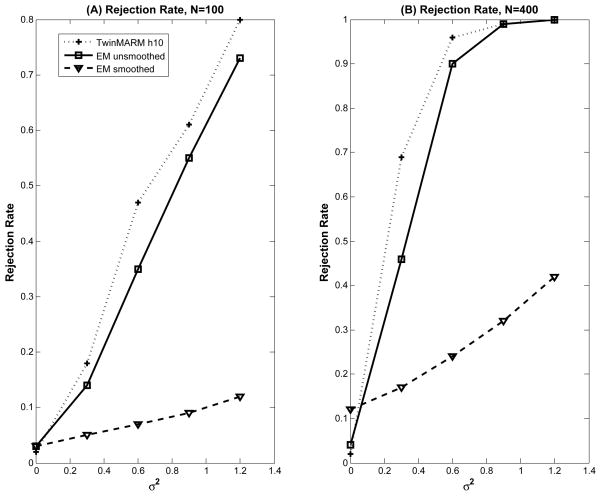
To assess both Type I and II error rates of WA(v; h) at the pixel level, we tested the hypotheses and across all pixels. We applied the same PS procedure and computed the p-values of WA(v; h) at each scale. The 1,000 replications were used to calculate the estimates and standard errors of rejection rates with significance level α = 5%. For the test statistic WA(v; h), the Type I rejection rates in ROI with were relatively small for all scales, while the statistical power for rejecting the null hypothesis in ROIs with significantly increased with larger radius h (Fig. 7). For all ROIs with , WA(v; h10) outperforms the likelihood ratio test statistic (LRT) based on the EM algorithm without smoothing and EM algorithm with smoothing in terms of power (Fig. 7).
Fig. 7.
Simulation results for WA(v; h): rejection rates for pixels inside the five ROIs were reported by using TwinMARM at the h10 scale, EM, EM after smoothing the same simulated data and 2 different sample sizes (n = 100, 400) at α = 5%. For each case, 1,000 simulated datasets were used.
Finally, we note that there are at least three limitations associated with our simulation studies. First, it should be noted that the simulated phantom image is 2D, whereas most real neuroimaging data is 3D. TwinMARM should work better in 3D and the variance reduction should be larger, since more neighboring voxels are available at each voxel for calculating adaptive weights and parameter estimation. Secondly, we did not directly simulate ‘real’ functional (or structural) imaging data from twin studies, since it is not straightforward to simulate real imaging data according to model (1). Thirdly, we did not evaluate the reconstruction and registration errors in reconstructing and registering real imaging data from twin studies.
4 Real-World Example
We considered the early postnatal brain development project led by Dr. Gilmore at the University of North Carolina at Chapel Hill [9]. This study was approved by the Institutional Review Board. A total of 49 paired twins (36 males and 62 females) were selected. All 49 pairs were scanned as neonates within a few weeks after birth at term. Written consent was obtained from their parents before imaging acquisition. The mean gestational age at MR scanning was 246 ± 18.3 days (range: 192 to 270 days). All infants were fed and calmed to sleep on a warm blanket with proper ear protection and they slept comfortably inside the MR scanner. None of infants was sedated during the imaging session.
We used a 3T Allegra head-only MR system (Siemens Medical Inc., Erlangen, Germany) to acquire all the images. The system was equipped with a maximal gradient strength of 40 mT/m and a maximal slew rate of 400 mT/(m·msec). We used a single shot EPI DTI sequence (TR/TE=5400/73 msec) with eddy current compensation to acquire the DTI images. The acquisition sequence consists of five repetitions of six gradient scans (b = 1000 s/mm2) at 6 non-collinear directions and a single reference scan (in total 5×7 = 35 images). The voxel resolution was isotropic 2 mm, and the in-plane field of view was set at 256 mm in both directions.
We then employed a nonlinear fluid deformation based high-dimensional, unbiased atlas computation method to process all 98 DTI datasets [58]. The atlas building procedure started with an affine registration and was followed by a nonlinear registration of a set of feature images for all subjects. The feature images are the maximum eigenvalue of the Hessian of the fractional anisotropy (FA) image, which are sensitive to the geometry of white matter. Using the computed deformation fields, we warped all tensor images into the unbiased atlas space via log-euclidean based interpolation [59]. We also averaged all the warped tensor images to create a study specific DTI atlas.
FA has been widely used as a measurement to assess directional organization of the brain which is greatly influenced by the magnitude and orientation of white matter tracts. Here, FA images are employed to identify the spatial patterns of white matter maturation. We considered a linear model yij(v) = β1(v) + β2(v)Gij + β3(v)Zij + aij(v) + ci(v) + εij(v) for i = 1, ···, 49 and j = 1, 2 at each voxel of the template, where Gij and Zij, respectively, represent the dummy variables for gender (male=1 and female=0) and zygote (MZ=1 and DZ=0), and aij(v), ci(v) and eij(v) are, respectively, the additive genetic, common environmental, and residual effects on the i-th twin pair as specified in Fig. 1. We focused on the major white matter regions. We calculated the MLE of parameters by using the EM algorithm and then applied our TwinMARM method with ch = 1.15 and S = 10 to carry out statistical analysis.
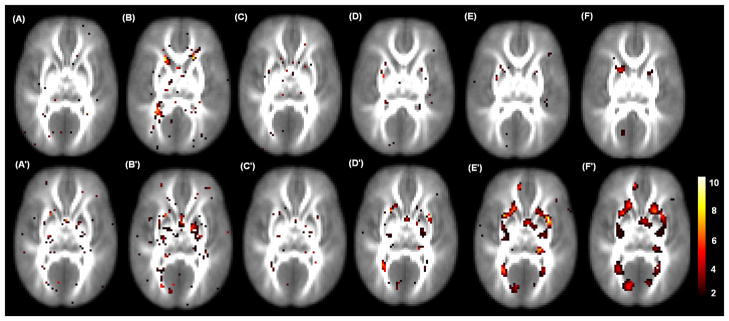
We tested for additive genetic effect across all voxels v in the white matter regions. To correct for multiple comparisons, we used the raw p-value smaller than 0.05 along with a 20 voxel extent threshold. We found significant regions in the inferior frontal gyrus, triangular part and mid cingulate cortex regions. We identified more voxels by using TwinMARM compared to EM (Table 1). We also tested for common environmental effect across all voxels v. Several interesting regions identified included the right precentral gyrus, inferior frontal gyrus, triangular part, supplementary motor area, insula, hippocampus, right fusiform, and thalamus. Fig. 8 shows some selected slices of – log10(p) map for environmental effect obtained from the 10th iteration of TwinMARM. The results show that TwinMARM may have significantly improved sensitivity and reliability as areas of significance in the TwinMARM results appear larger and smoother compared to the voxel-wise analysis approach, which is close to the result obtained from the first iteration of TwinMARM. The FA measure shows strong effects of genetic factors and common environment in the frontal lobes and in the limbic lobes. The detection of a genetic effect is partially consistent with many other studies that have detected strong genetic influences on brain morphology [11, 4, 60, 55].
Table 1.
Real data analysis results. # of voxels whose p–value is less than a specific threshold for testing the genetic and environmental effect for EM method without smoothing, smoothing with Gaussian kernels with FWHM equal to 3mm, 6mm, and 9mm and TwinMARM method at the 1st and 10th iteration.
| EM smoothness (mm) | TwinMARM (round) | ||||||
|---|---|---|---|---|---|---|---|
| Effect | 0 | 3 | 6 | 9 | 1st | 10th | |
| p–value< 0.01 | Genetic | 13 | 26 | 19 | 32 | 19 | 47 |
| Environment | 11 | 41 | 140 | 152 | 23 | 47 | |
|
| |||||||
| p–value< 0.05 | Genetic | 148 | 233 | 208 | 254 | 198 | 433 |
| Environment | 171 | 567 | 1318 | 1984 | 232 | 715 | |
|
| |||||||
| p–value< 0.10 | Genetic | 548 | 865 | 812 | 719 | 676 | 1430 |
| Environment | 615 | 1708 | 3769 | 5804 | 956 | 2568 | |
Fig. 8.
Results from the 49 twin pairs in a neonatal project on brain development on the selected 27th and 30th slices. Panels (A)–(F) : the – log10(p) values for testing genetic effects by using TwinMARM at the 1st and 10th iterations and EM with FWHM equal to 0mm, 3mm, 6mm, 9mm for the 27th slice; Panels (A′)–(F′) : the corresponding – log10(p) values for testing environmental effects for the 30th slice.
We also observed the smoother and larger significant clusters of genetic factors and common environment by using the EM algorithm with smoothed data from FWHM=6mm and 9mm (Fig. 8(F), (E′) and (F′)). First, based on our simulation studies, we observed that the EM algorithm for unsmoothed data and TwinMARM do not introduce bias in the estimates of , whereas we observed the substantial bias in the estimates of for the heavily smoothing imaging data (Figs. 3 and 5). Secondly, these smoother and larger significant clusters do not appear in the results obtained from the unsmoothed data. Thus, we feel that these smoother and larger significant clusters obtained from the heavily smoothed data may be false positive results (Fig. 8 and Table 1 and 2). However, a word of caution is needed in interpreting these significant clusters to be false positive, since two key assumptions underlying EM and TwinMARM, including the perfect registration assumption and the structural equation model (1), may be invalid for this real data set.
Table 2.
Real data analysis results. # of cluster which has over 20 contiguous voxels with p–value less than a specific threshold for testing the genetic and environmental effect by using EM method without smoothing, smoothing with Gaussian kernels (FWHM=3mm, 6mm and 9mm) and TwinMARM method at the 1st and 10th iteration.
| EM smoothness (mm) | TwinMARM (round) | ||||||
|---|---|---|---|---|---|---|---|
| Effect | 0 | 3 | 6 | 9 | 1st | 10th | |
| p–value< 0.01 | Genetic | 0 | 0 | 0 | 1 | 0 | 0 |
| Environment | 0 | 0 | 1 | 2 | 0 | 0 | |
|
| |||||||
| p–value< 0.05 | Genetic | 0 | 0 | 3 | 3 | 0 | 3 |
| Environment | 0 | 4 | 17 | 11 | 0 | 4 | |
|
| |||||||
| p–value< 0.10 | Genetic | 0 | 10 | 9 | 8 | 1 | 9 |
| Environment | 4 | 21 | 18 | 10 | 3 | 20 | |
5 Conclusions and Discussion
We have developed TwinMARM for the statistical analysis of twin neuroimaging data. TwinMARM consists of two stages of multiscale adaptive regression methods. TwinMARM have three features: being spatial, being hierarchical, and being adaptive. The TwinMARM procedure is a simultaneous smoothing and estimation procedure. Each stage of TwinMARM constructs hierarchical nested spheres with increasing radii at all voxels to adaptively compute weighted quasi-likelihood functions and efficiently utilizes available information to obtain parameter estimates. This leads to improved accuracy of parameter estimates and increased statistical power in detecting subtle changes of brain structure and function.
As shown in the simulation studies, the commonly used Gaussian kernel for smoothing imaging data can introduce two levels of biases for the analysis of twin imaging data, which can dramatically increase the numbers of false positives and negatives. The first level of biases originates from the mean structure, which is associated with the regression coefficients β(v). Most smoothing methods, which are independent of imaging data, usually blur the image data near the edges of the significant regions, and consequently introduce biases in β(v). The second level of biases comes from the variance structure, which is associated with the genetic coefficients σ2(v). Most smoothing methods are not only conducted independently of imaging data but also of the statistical model fitted to the imaging data. Statistically, standard smoothing methods can change the variance structure in the imaging data which are the focus of twin imaging studies. Thus, the second level of biases is much more severe than the first level of biases. In practice, we should avoid directly smoothing twin imaging data.
Several limitations need to be addressed in future research. The TwinMARM procedure is based on the two key assumptions, including the perfect registration assumption and the structural equation model (1). A first limitation of our study is that the perfect registration assumption is demonstrably false for real imaging data from twin studies. It is important and interesting to integrate the registration method, smoothing method, and TwinMARM into a single framework in order to appropriately account for registration errors in the statistical analysis of imaging data from twin studies. A second limitation of our study is that the structural equation model (1) can be invalid for real imaging data from twin studies. Heritability estimates used for most twin studies rest on restrictive assumptions in model (1), which are usually not tested, and if they are, can often found to be violated by the real data [61].
There are several current topics for our research. We are developing a Graphical User Interface (GUI) to pack our code for TwinMARM, which will be freely downloadable from our website at http://www.bios.unc.edu/research/bias. The development of multiscale-adaptive methods to determine multiscale neighborhoods that adapt to the pattern of imaging data at each voxel would undoubtedly be a useful resource for neuroimaging researchers. Further, we will extend TwinMARM from the univariate to multivariate and complex imaging measures, such as deformation tensors. We will develop a multiscale adaptive model for neuroimaging data from familial studies [25]. More research is needed for optimizing the choices of the parameters in TwinMARM. It is interesting to incorporate other edge-preserving local smoothing methods into TwinMARM and MARM [33, 34, 37, 38].
6 Appendix: Effect of smoothing twin imaging data
We examined the effect of smoothing twin imaging data on its associated statistical analysis as follows. As an illustration, we focus on an arbitrary voxel v and its spherical neighborhood, denoted by {v′ : v′ ∈ B(v, h)}. Standard smoothing methods primarily take a weighted average of all observations {yij(v′) : v′ ∈ B(v, h)}, denoted by ỹij(v), such that ỹij(v) = ∑v′ ∈ B(v,h) ω(v, v ′; h)yij(v′), where ∑v′ ∈ B(v,h) ω(v, v′; h) = 1 and ω(v, v; h) > 0. Under model (1), we can show that
| (22) |
where β̃(v) = ∑v′∈B(v,h) ω(v, v′; h)β(v′),
It can be shown that the variance of ãij(v), denoted by , depends on both the variances of {aij(v′) : v′ ∈ B(v, h)} and their spatial correlations. Specifically, we have
Similar arguments hold for the variances of d̃ij(v), c̃ij(v) and ẽij(v) denoted by , and , respectively. Due to the presence of spatial correlations and other factors (e.g., the inhomogeneity of ) across v′ ∈ B(v, h)), it is common that
Generally, the Gaussian kernel smoothing method can easily modify the variance structure of twin imaging data. Because such variance structures are often the focus of twin studies, we thus suggest avoiding directly smoothing twin imaging data in practice.
We show that TwinMARM does not introduce bias in the variance structure of twin imaging data. This consists of two steps.
Step (i) As n → ∞, β̂(v; hS) converges β*(v) in probability and is asymptotically normal distributed for all v ∈
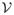 under the mild conditions given in [43], where β*(v) is the true value β(v) in voxel v. Please see Theorems 1–3 in [43] for the details.
under the mild conditions given in [43], where β*(v) is the true value β(v) in voxel v. Please see Theorems 1–3 in [43] for the details.Step (ii) As n → ∞, η̃(v; hS) converges η*(v) and converges to a random variable in distribution for all v ∈
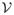 . We can essentially use the same idea in proving Step (i) to prove Step (ii), but there is a complexity in dealing with substituting β*(v) by β̂(v;hS). We will show that such substitution is negligible below.
. We can essentially use the same idea in proving Step (i) to prove Step (ii), but there is a complexity in dealing with substituting β*(v) by β̂(v;hS). We will show that such substitution is negligible below.
We just highlight the key idea underlying the proof of Step (ii). It follows from Step (i) and the ACE model that for j = 1, 2,
| (23) |
Let Δβ(v; hS) = {β̂(v; hS) – β*(v)}. With some simple algebra, for j = 1, 2, we can show that
in which we have used the results in Step (i) and (van der Vaart, 1998) and set
| (24) |
Similarly, we can show that
| (25) |
where . Based on these results, we can obtain model (14), in which εi(v) is almost unbiased except an Op(n−1/2) term. We will show that such Op(n−1/2) term has a negligible effect below.
Then, we need to show that η̂(v; h) converges η*(v) in probability and
asymptotically converges a random variable in distribution for all v ∈
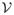 under some mild conditions, which are similar to those give in [43]. We focus on h = h0 = 0. For h > 0, we can generalize the proofs given in [43]. Recall that
. We define
under some mild conditions, which are similar to those give in [43]. We focus on h = h0 = 0. For h > 0, we can generalize the proofs given in [43]. Recall that
. We define
| (26) |
By using the law of large number [62], we have
in which we have used obtained from Step (i) and converges to zero in probability. Thus, it follows from (14) that
which yields that it is negligible to substitute β*(v) by β̂(v; hS). Furthermore, we calculate
where . The choice of ωe(v, v′; h) ensures the convergence of η̂(v; h1). Specifically, if η*(v) ≠ η*(v′), then we have Dη(v, v′; h) = Op(n1/2) and Kst(Dη(v, v′; h)/Cn) converges to infinity and zero, respectively. Therefore, ωe(v, v′; h) can give a small weight to voxel v′. Similar to the proof of Theorems 1–3 in [43], we can show that as n → ∞, η̂(v; h) converges η*(v) and . Finally, we can prove Step (ii) for any fixed S.
Footnotes
This work was supported in part by NIH grants RR025747-01, P01CA142538-01, MH086633, and AG033387 to Dr. Zhu, NIH grants MH064065, HD053000, and MH070890 to Dr. Gilmore, NIH grants R01NS055754 and R01EB5-34816 to Dr. Lin, Lilly Research Laboratories, the UNC NDRC HD 03110, Eli Lilly grant F1D-MC-X252, and NIH Roadmap Grant U54 EB005149-01, NAMIC to Dr. Styner. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The readers are welcome to request reprints from Dr. Hongtu Zhu,, hzhu@bios.unc.edu; phone: 919-966-7272.
References
- 1.Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- 2.Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. NeuroImage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- 3.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: a review of brain imaging studies in twins. Human Brain Mapping. 2007;28:446–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang MC, Barysheva M, Shattuck DW, Lee AD, Madsen SK, Avedissian C, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Wright MJ, Srivastava A, Balov N, Thompson PM. Genetics of brain fiber architecture and intellectual performance. Journal of Neuroscience. 2009;29:2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon U, Fahim C, Perusse D, Evans AC. Lateralized genetic and environmental influences on human brain morphology of 8-year-old twins. NeuroImage. 2010;53:1117–1125. doi: 10.1016/j.neuroimage.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pell GS, Briellmann RS, Lawrence KM, Glencross D, Wellard RM, Berkovic SF, Jackson GD. Reduced variance in monozygous twins for multiple mr parameters: Implications for disease studies and the genetic basis of brain structure. NeuroImage. 2010;49:1536–1544. doi: 10.1016/j.neuroimage.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Jahanshad N, Lee AD, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM. Genetic influences on brain asymmetry: A dti study of 374 twins and siblings. NeuroImage. 2010;52:455–469. doi: 10.1016/j.neuroimage.2010.04.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou YY, Leporé N, Chiang MC, Avedissian C, Barysheva M, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Toga AW, Thompson PM. Mapping genetic influences on ventricular structure in twins. Neuroimage. 2009;44:1312– 1323. doi: 10.1016/j.neuroimage.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmore JH, Schmitt JE, Knickmeyer RA, Smithm JK, Lin W, Styner M, Gerig G, Neale MC. Genetic and environmental contributions to neonatal brain structure: a twin study. Human Brain Mapping. 2010;31:1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci USA. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen V, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga A. Genetic influences on brain structure. Nature Neuroscience. 2001;4:1253–1358. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 12.Yue Y, Loh JM, Lindquist MA. Adaptive spatial smoothing of fmri images. Statistics and its Interface. 2010;3:3–14. [Google Scholar]
- 13.Kempthorne O, Osborne RH. The interpretation of twin data. American Journal of Human Genetics. 1961;13:320–339. [PMC free article] [PubMed] [Google Scholar]
- 14.Haseman JK, Elston RC. The estimation of genetic variance from twin data. Behavior Genetics. 1970;1:11–19. doi: 10.1007/BF01067367. [DOI] [PubMed] [Google Scholar]
- 15.Neale MC, Heath AC, Hewitt JK, Eaves LJ, Fulker DW. Fitting genetic models with lisrel: Hypothesis testing. Behavior Genetics. 1989;19:37–49. doi: 10.1007/BF01065882. [DOI] [PubMed] [Google Scholar]
- 16.Feng R, Zhou G, Zhang M, Zhang H. Analysis of twin data using sas. Biometrics. 2009;65:584–589. doi: 10.1111/j.1541-0420.2008.01098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Ser B. 1995;57:289–300. [Google Scholar]
- 19.Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003;12:419–426. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 20.Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J. Unified univariate and multivariate random field theory. NeuroImage. 2004;23:189–195. doi: 10.1016/j.neuroimage.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 21.Chumbley J, Worsley J, Flandin KG, Friston KJ. Topological fdr for neuroimaging. Neuroimage. 2010;49(4):3057–3064. doi: 10.1016/j.neuroimage.2009.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salimi-Khorshidi G, Smith SM, Nichols TE. Adjusting the effect of nonstationarity in cluster-based and tfce inference. Neuroimage. 2011;54(3):2006–2019. doi: 10.1016/j.neuroimage.2010.09.088. [DOI] [PubMed] [Google Scholar]
- 23.Hecke WV, Sijbers J, Backer SD, Poot D, Parizel PM, Leemans A. On the construction of a ground truth framework for evaluating voxel-based diffusion tensor mri analysis methods. NeuroImage. 2009;46:692–707. doi: 10.1016/j.neuroimage.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 24.Jones DK, Symms MR, Cercignani M, Howard RJ. The effect of filter size on vbm analyses of dt-mri data. NeuroImage. 2005;26:546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Zhu HT, Li YM, Tang NS, Bansal R, Hao XJ, Weissman MM, Peterson BG. Statistical modelling of brain morphological measures within family pedigrees. Statistica Sinica. 2008;18:1554–1569. [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang MC, Avedissian C, Barysheva M, Toga AW, McMahon KL, Zubicaray GI, Wright MJ, Thompson PM. Extending genetic linkage analysis to diffusion tensor images to map single gene effects on brain fiber architecture. MICCAI ‘09 Proceedings of the 12th International Conference on Medical Image Computing and Computer-Assisted Intervention: Part II; 2009. pp. 506–513. [DOI] [PubMed] [Google Scholar]
- 27.Besag JE. On the statistical analysis of dirty pictures (with discussion) Journal of the Royal Statistical Society, Ser B. 1986;48:259–302. [Google Scholar]
- 28.Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. CRC/Chapman and Hall; 2004. [Google Scholar]
- 29.Penny WD, Trujillo-Barreto NJ, Friston KJ. Bayesian fmri time series analysis with spatial priors. NeuroImage. 2005;24:350–362. doi: 10.1016/j.neuroimage.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Groves AR, Chappell MA, Woolrich MW. Combined spatial and non-spatial prior for inference on mri time-series. NeuroImage. 2009;45(3):795–809. doi: 10.1016/j.neuroimage.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 31.Zhu HT, Gu M, Peterson BS. Maximum likelihood from spatial random effects models via the stochastic approximation expectation maximization algorithm. Statistics and Computing. 2007;15:163–177. [Google Scholar]
- 32.Bowman FD. Spatio-temporal models for region of interest analyses of functional mappping experiments. Journal of American Statistical Association. 2007;102:442–453. [Google Scholar]
- 33.Qiu P. Image Processing and Jump Regression Analysis. New York: John Wiley & Sons; 2005. [Google Scholar]
- 34.Qiu P. Jump surface estimation, edge detection, and image restoration. Journal of American Statistical Association. 2007;102:745–756. [Google Scholar]
- 35.Polzehl J, Spokoiny VG. Adaptive weights smoothing with applications to image restoration. J R Statist Soc B. 2000;62:335–354. [Google Scholar]
- 36.Polzehl J, Spokoiny VG. Propagation-separation approach for local likelihood estimation. Probab Theory Relat Fields. 2006;135:335–362. [Google Scholar]
- 37.Qiu P, Mukherjee PS. Edge structure preserving image denoising. Signal Processing. 2010;90:2851–2862. [Google Scholar]
- 38.Mukherjee PS, Qiu P. 3-d 3-d image denoising by local smoothing and nonparametric regression. Technometrics. 2011;53:196–208. [Google Scholar]
- 39.Tabelow K, Polzehl J, Voss HU, Spokoiny V. Analyzing fmri experiments with structural adaptive smoothing procedures. NeuroImage. 2006;33:55–62. doi: 10.1016/j.neuroimage.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Tabelow K, Polzehl J, Spokoiny V, Voss HU. Diffusion tensor imaging: structural adaptive smoothing. NeuroImage. 2008;39:1763–1773. doi: 10.1016/j.neuroimage.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 41.Tabelow K, Polzehl J, Ulug AM, Dyke JP, Watts R, Heier LA, Voss HU. Accurate localization of brain activity in presurgical fmri by structure adaptive smoothing. IEEE Trans Med Imaging. 2008;27:531–537. doi: 10.1109/TMI.2007.908684. [DOI] [PubMed] [Google Scholar]
- 42.Polzehl J, Voss HU, Tabelow K. Structural adaptive segmentation for statistical parametric mapping. Neuroimage. 2010;52:515–523. doi: 10.1016/j.neuroimage.2010.04.241. [DOI] [PubMed] [Google Scholar]
- 43.Li YM, Zhu HT, Shen DG, Lin WL, Gilmore J, Ibrahim JG. Multiscale adaptive regression models for neuroimaging data. Journal of the Royal Statistical Society, Series B. 2011 doi: 10.1111/j.1467-9868.2010.00767.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner J, Friston KJ. Voxel-based morphometry: the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 45.Chung MK, Dalton KM, Davidson RJ. Tensor-based cortical surface morphometry via weighted spherical harmonic representation. IEEE Trans Med Imaging. 2008;27:1143– 1151. doi: 10.1109/TMI.2008.918338. [DOI] [PubMed] [Google Scholar]
- 46.Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci USA. 2005;102:4872–4677. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson PM, Toga AW. A framework for computational anatomy. Comput Visual. 2002;5:13–34. [Google Scholar]
- 48.Plomin R, Kosslyn SM. Genes, brain and cognition. Nature Neuroscience. 20011;4:1153–1154. doi: 10.1038/nn1201-1153. [DOI] [PubMed] [Google Scholar]
- 49.Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating population-based brain atlases. Human Brain Mapping. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu HT, Ibrahim JG, Tang N, Rowe DB, Hao X, Bansal R, Peterson BS. A statistical analysis of brain morphology using wild bootstrapping. IEEE Trans Med Imaging. 2007;26:954–966. doi: 10.1109/TMI.2007.897396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Guo X, He M, Zhang H. Statistical inference in mixed models and analysis of twin and family data. Biometrics. 2011 doi: 10.1111/j.1541-0420.2010.01548.x. page in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman: Harlow, Essex; 1996. [Google Scholar]
- 53.Polzehl J, Spokoiny VG. Image denoising: pointwise adaptive approach. Annals of Statistics. 2003;31:30–57. [Google Scholar]
- 54.Meyer MC. An algorithm for quadratic programming with applications in statistics. 2009. [Google Scholar]
- 55.Thompson PM, Cannon TD, Toga AW. Mapping genetic influences on human brain structure. Ann Med. 2002;24:523–536. doi: 10.1080/078538902321117733. [DOI] [PubMed] [Google Scholar]
- 56.Shapiro A. Towards a unified theory of inequality constrained testing in multivariate analysis. International Statistical Review. 1988;56:49–62. [Google Scholar]
- 57.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the em algorithm. Journal of the Royal Statistical Soceity Series B. 1977;39:1–38. [Google Scholar]
- 58.Goodlett CB, Fletcher PT, Gilmore JH, Gerig G. Group analysis of dti fiber tract statistics with application to neurodevelopment. NeuroImage. 2009;45:S133–S142. doi: 10.1016/j.neuroimage.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arsigny V, Fillard P, Pennec X, Ayache N. Log-euclidean metrics for fast and simple caculus on diffusion tensors. Magnetic Resonance in Medicine. 2006;56:411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- 60.Chiang MC, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM. Genetics of white matter development: A dti study of 705 twins and their siblings aged 12 to 29. NeuroImage. 2011;54:2308–2317. doi: 10.1016/j.neuroimage.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schonemann P. Models and muddles of heritability. Genetica. 1997;99:97–108. doi: 10.1007/BF02259513. [DOI] [PubMed] [Google Scholar]
- 62.van der Vaar AW. Asymptotic Statistics. Cambridge University Press; 1998. [Google Scholar]