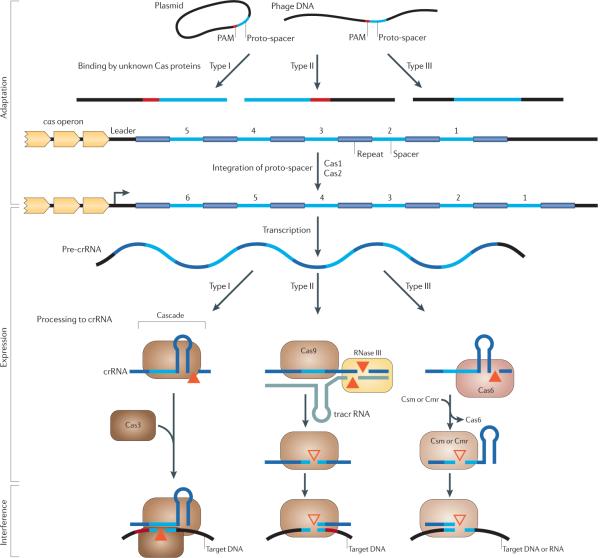
Figure 1. The three stages of CRISPR–Cas action.
CRISPR-Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems act in three stages: adaptation, expression and interference. In type I and type II CRISPR-Cas systems, but not in type III systems, the selection of proto-spacers in invading nucleic acid probably depends on a proto-spacer-adjacent motif (PAM)22,30,31, but how the PAM or the nucleic acid is recognized is still unclear. After the initial recognition step, Cas1 and Cas2 most probably incorporate the proto-spacers into the CRISPR locus to form spacers. During the expression stage, the CRISPR locus containing the spacers is expressed, producing a long primary CRISPR transcript (the precrRNA). The CRISPR-associated complex for antiviral defence (Cascade) complex binds the pre-crRNA, which is then cleaved by the Cas6e or Cas6f subunits (in subtype I-E or I-F, respectively), resulting in crRNAs with a typical 8-nucleotide repeat fragment on the 5′ end and the remainder of the repeat fragment, which generally forms a hairpin structure, on the 3′ flank. Type II systems use a trans-encoded small RNA (tracrRNA) that pairs with the repeat fragment of the pre-crRNA, followed by cleavage within the repeats by the housekeeping RNase III in the presence of Cas9 (formerly known as Csn1 or Csx12). Subsequent maturation might occur by cleavage at a fixed distance within the spacers25, probably catalysed by Cas9. In type III systems, Cas6 is responsible for the processing step, but the crRNAs seem to be transferred to a distinct Cas complex (called Csm in subtype III-A systems and Cmr in subtype III-B systems). In subtype III-B systems, the 3′ end of the crRNA is trimmed further28. During the interference step, the invading nucleic acid is cleaved. In type I systems, the crRNA guides the Cascade complex to targets that contain the complementary DNA, and the Cas3 subunit is probably responsible for cleaving the invading DNA21. The PAM probably also plays an important part in target recognition in type I systems. In type II and type III systems, no Cas3 orthologue is involved (TABLE 2). In type II systems, Cas9 loaded with crRNA probably directly targets invading DNA, in a process that requires the PAM26. The two subtypes of CRISPR–Cas type III systems target either DNA (subtype III-A systems31) or RNA (subtype III-B systems28). In type III systems, a chromosomal CRISPR locus and an invading DNA fragment are distinguished by either base pairing to the 5′ repeat fragment of the mature crRNA (resulting in no interference) or no base pairing (resulting in interference)30. Filled triangles represent experimentally characterized nucleases, and unfilled triangles represent nucleases that have not yet been identified.