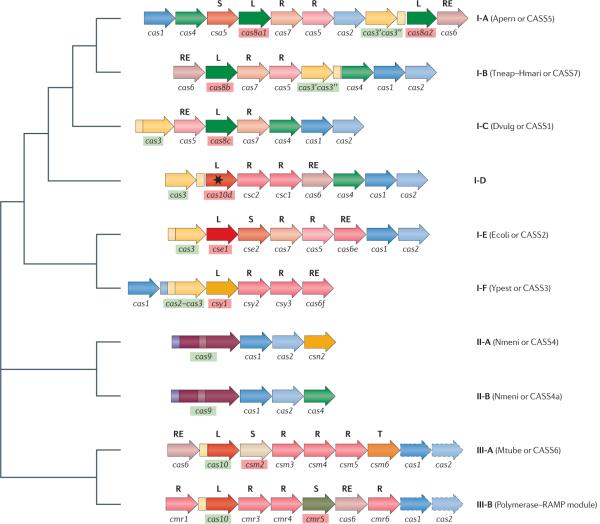
Figure 2. The relationship of the three major types and ten sub-types of CRISPR systems.
The typical, simplest operon architectures are shown for each type and subtype of CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) system; numerous variations exist. Orthologous genes are colour coded and identified by a family name, as given in TABLE 2. The signature genes for CRISPR–Cas types are shown within green boxes, and those for sub-types are shown within red boxes. The letters above the genes show major categories of Cas proteins: large CRISPR-associated complex for antiviral defence (Cascade) subunits (L), small Cascade subunits (S), repeat-associated mysterious protein (RAMP) Cascade subunits (R), RAMP family RNases involved in crRNA processing (RE) (note that only those in subtypes I-E, I-F and III-B systems have been characterized), and transcriptional regulators (T). The star indicates a predicted inactivated polymerase with an HD domain. For subtype I-A systems, the cas8a1 and cas8a2 genes are typically mutually exclusive but both can be considered signature genes for the subtype. For type III systems, the cas1 and cas2 genes in dashed boxes are not associated with all type III polymerase–RAMP modules. In addition to previously published data, this schematic shows Cas7 (COG1857) as a member of the RAMP superfamily. For each CRISPR–Cas subtype (except for the newly identified subtype I-D), the old names from REFS 13,14 are indicated in parentheses. Figure is modified, with permission, from REF. 14 © (2006) BioMed Central.