Figure 3. Phylogenetic tree for Cas1 (COG1518) proteins.
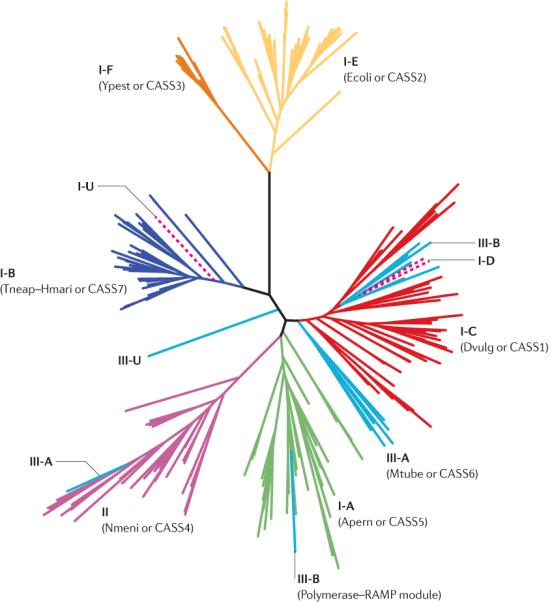
The BLASTCLUST program was used to cluster the sequences of CRISPR (clustered regularly interspaced short palindromic repeats)-associated protein 1 (Cas1) by similarity (parameters: the sequence length to be covered was 75%, and the score identity threshold was 0.9), and one representative from each cluster was chosen (see the list in Supplementary information S4 (table)). Six major subtypes of type I CRISPR–Cas system (I-A to I-F), as well as type II and type III systems, are colour coded. Dashed lines show cas1 genes that are found in `hybrid' CRISPR loci containing genes from both type I and type III CRISPR–Cas systems (see main text for details). Subtypes I-U and III-U (U for unclassified) denote CRISPR–Cas systems that lack currently defined subtype-specific signature genes (see main text for details). The maximum likelihood tree was constructed using the PHYML program46, from 182 informative positions in the multiple alignment of a representative set of 228 Cas1 proteins from 442 complete genomes (those that encode Cas1 from the set of 703 genomes listed in Supplementary information S1 (table)). For each CRISPRCas subtype (except for the newly identified subtype I-D), the old names from REFS 13,14 are indicated in parentheses.
