Fig. 3.
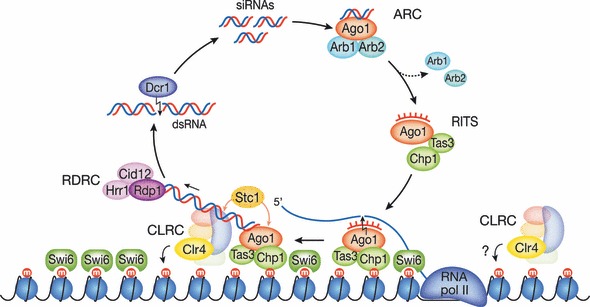
Schematic model of the self-enforcing loop for RNAi-mediated epigenetic silencing in Schizosaccharomyces pombe. Heterochromatin is shown as DNA (black line) wrapped around nucleosomes (blue circles) modified by histone H3 lysine 9 methylation (H3K9me; red circles “m”). Nascent transcripts generated by RNA polymerase II are targeted by an activated RNAi-induced transcriptional silencing (RITS) complex that also interacts with H3K9me through the Chp1 subunit. RDRC is then recruited for synthesis of dsRNA from the transcripts, which is cleaved into duplex siRNAs by Dcr1 and bound by Argonaute siRNA chaperone (ARC). These siRNAs are then processed into single-stranded siRNAs and loaded back into RITS completing the loop. Catalytically active RITS recruits the CLR4 complex (CLRC) for spreading of epigenetic silencing into adjacent regions through interaction with Stc1, although CLRC can also associate with these regions independent of RNAi and recruit inactive RITS.
