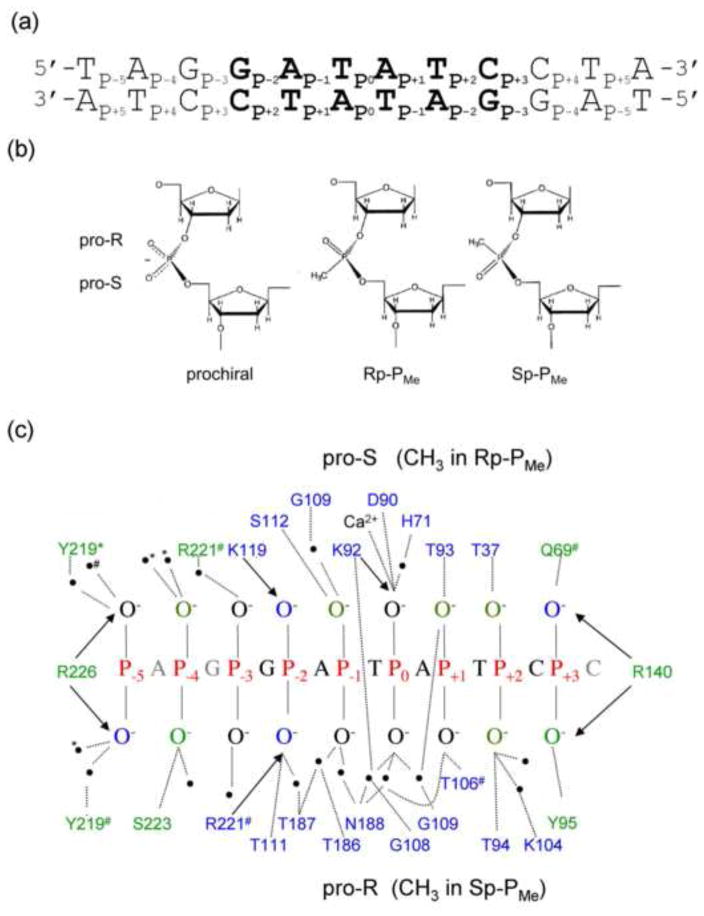
Figure 2. Phosphate contacts made in the specific EcoRV-DNA recognition complex.
(a) Specific DNA substrate used for all thermodynamic experiments. The specific recognition sequence GATATC (bold) is embedded in a 24 bp parent oligonucleotide. All PMe substitutions were made symmetrically in both DNA strands. (b) Comparison of anionic prochiral phosphodiester (left) with uncharged chiral methylphosphonates (PMe). By IUPAC convention, replacement of the pro-S (O1P) non-bridging phosphoryl oxygen with a methyl group results in an Rp-PMe diastereomer; replacement of the pro-R (O2P) phosphoryl oxygen results in an Sp-PMe diastereomer. (c) Phosphate contacts made to one strand of the specific EcoRV-bound DNA, based on 10 crystal structures of EcoRV-DNA ternary complexes. The base sequence is that used in our thermodynamic experiments. Hydrogen bonds (<3.5Å) to the phosphoryl oxygens from polar side chains and water molecules (black circles) are shown as dotted lines; Coulombic interactions (≤4.3Å) with cationic sidechains are denoted by arrows. Most contacts are symmetrical with respect to protein subunits and DNA strands; those contacts made to only one DNA chain are denoted by # or *; these correspond to chains C and D in PDB ID: 1B94. Sidechains that contact phosphoryl oxygens within the GATATC recognition site are in blue; those making contact outside the GATATC site are in green.