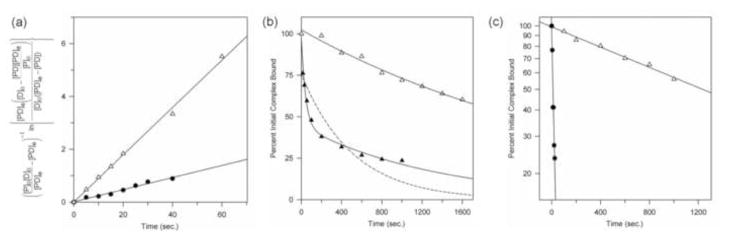
Figure 8. Effect of PMe on association and dissociation of K119A-DNA complexes.
(a) Representative kinetics of association of K119A mutant EcoRV (1 nM) with (△) unmodified DNA (0.5 nM) or (●) RP-PMe-2 DNA (0.5 nM) determined in binding buffer plus 0.14M K+. The ordinate is the integrated-rate expression for a reversible bimolecular process. The slope of the curve (△, R2 = 0.95; ●, R2 = 0.98) is the apparent association rate constant ka.
(b) Representative kinetics of dissociation of complexes of K119A mutant EcoRV with (△) RP-PMe-2 DNA or (▲) racemic RP,SP-PMe-2 DNA at 0.1M K+. (Lower salt is required to detect the faster rate component for racemic DNA.) K119A-RP-PMe-2 complexes were formed at 1.9 nM DNA, 1.5 nM enzyme (compare KD = 0.38 pM) and measurement initiated with 7.6 μM cold competitor DNA. Racemic complex data at 2.7 nM DNA, 2.2. nM enzyme (compare KD = 1.1 pM), 2.7 μM competitor DNA. The ordinate is a linear scale. For the K119A-RP-PMe-2 complex, R2 =0.98 for both single and double exponential fits. For the racemic DNA, the dashed line shows the best single-exponential fit (R2 =0.78); the solid line shows a double-exponential fit (R2 =0.99). The biphasic curve for racemic DNA implies that complexes with RP-PMe-2 DNA are much more stable than those with SP-PMe-2 DNA, because the latter substitution disrupts stabilizing interactions with R221 and T111.
c) Semilog plot of the dissociation of K119A mutant EcoRV complexes with (●) unmodified DNA and (△) RP-PMe-2 DNA at 0.14M K+. The relief of electrostatic repulsion by the PMe-2 markedly stabilizes the complex against dissociation. Unmodified complex data at 0.43 μM DNA, 0.34 μM enzyme (compare KD = 1.7 nM), 43 μM competitor. RP-PMe-2 complex data at 7.8 nM DNA, 6.3 nM enzyme (compare KD = 16 pM), 7.8 μM competitor.