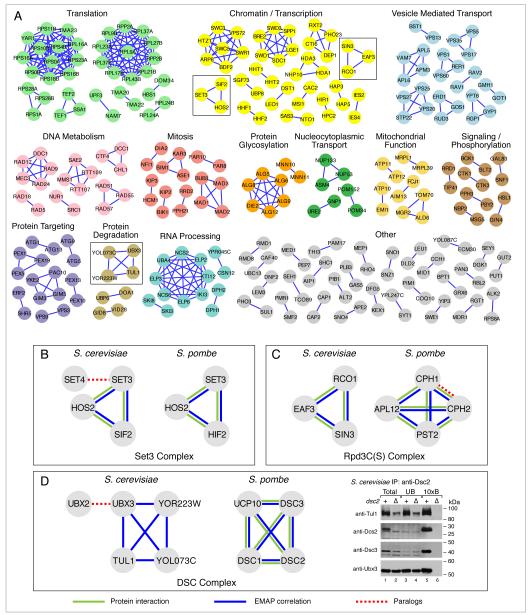
Figure 4. Conserved Functional Modules.
(A) Groups of genes with highly correlated genetic interaction profiles in both S. pombe and S. cerevisae are shown. S. cerevisiae gene names were used for labeling, as many of the S. pombe orthologs lack common names. Modules are manually grouped and colored according to the biological process they are involved in. Modules from the insets are boxed and correspond to the Set3 complex (B), the Rpd3C(S) (C) and the DSC complex (D). A full list of the modules identified, and their S. pombe counterparts, is given in Table S2. Blue edges correspond to pairs of genes that have high E-MAP similarity scores, green edges represent pairs of factors that are physically associated from previous studies whereas dashed red edges represent paralogs within one species. For the immunoprecipitation assay in (D), Dsc2 binding proteins were immunopurified from detergent lysates of wild-type and dsc2Δ cells using anti-Dsc2 affinity purified polyclonal antibody. Equal amounts of total (lanes 1 and 2) and unbound fractions (lanes 3 and 4) along with 10× bound fractions (lanes 5 and 6) were immunoblotted using the indicated HRP-conjugated antibodies. See Figure S6 for an additional experiment confirming that Dsc2 binds to Ubx3, but not Ubx2.