Figure 1. Structures of the pre- and post-Cre floxed mouse Zip4 gene and integration and genotyping screen designs.
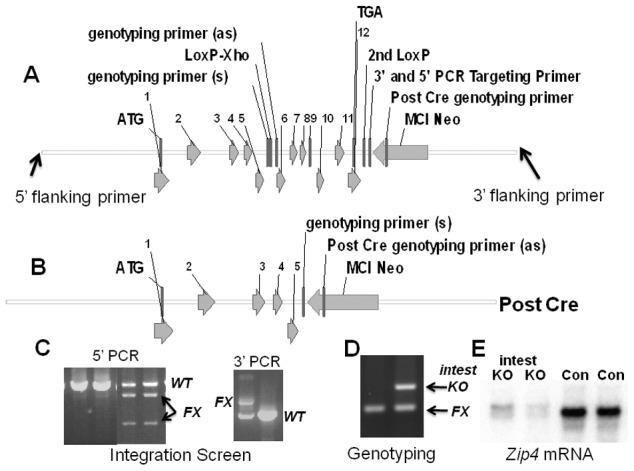
(A) The mouse Zip4 gene was captured using gap-repair and then manipulated using recombineering and the galK selection system. Exons (1–12) are indicated, as are the positions of LoxP sites (intron 5 and downstream of exon 12), the mc1-neomycin (MC1-Neo) cassette and the primers used to screen for integration and genotyping. (B) The structure of the Zip4 gene after Cre recombination is shown. The floxed Zip4 gene was targeted into E14 ES cells. (C) Properly targeted ES cells were identified by long range PCR using flanking and internal primers. PCR products from the control (Con) and floxed (FX) alleles are indicated. XhoI cleavage was used to differentiate between the floxed and wild-type alleles in the 5′ PCR screen whereas the 3′ PCR screen yielded the predicted larger product from the floxed allele. Targeted ES cells were used to generate mice homozygous for the floxed Zip4 allele. (D) Mice were genotyped by PCR amplification of the intron 5 region containing the LoxP site. The PCR product from homozygous mice before Cre-induced recombination is shown (left lane). A vil-CreERT2 transgene was bred into mice homozygous for the floxed Zip4 gene to allow for tamoxifen induction of Cre activity specifically in the intestinal epithelium. DNA from the intestine of tamoxifen injected mice was amplified by PCR to examine the extent of recombination of the floxed gene (right lane). PCR products from the recombined floxed intestine allele (intest KO) and the remaining floxed alleles (FX) shown. About 50% of the cells in the intestine are epithelial which is reflected in the extent of recombination shown. (E) Northern blot detection of intestine Zip4 mRNA in control (Con) and Zip4-intestine knockout mice (intest KO) 3 days after initiation of the knockout.
