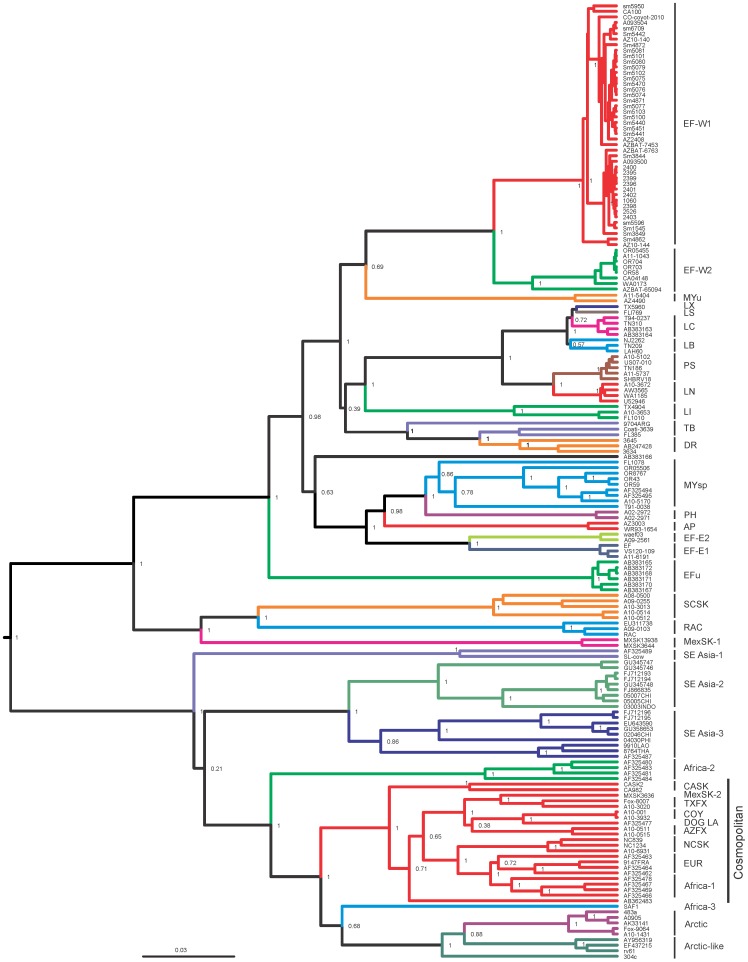
Figure 1. Bayesian tree of viruses, included in the study, based on the entire coding region of the G gene (1572 nuc).
Lineage abbreviations: EF-W1 and EF-W2 – Eptesicus fuscus, with predominantly western distribution; MYu – Myotis yumanensis; LX – Lasiurus xanthinus; LS – Lasiurus seminolus; LC – Lasiurus cinereus; LB – Lasiurus borealis; PS – Perimyotis subflavus; LN – Lasionycteris noctivagans; LI – Lasiurus intermedius; TB – Tadarida brasiliensis; DR – Desmodus rotundus; MYsp – Myotis spp; PH – Parastrellus hesperus; AP – Antrozous pallidus; EF-E1 and EF-E2 – Eptesicus fuscus, with predominantly eastern and central distribution; EFu – Eptesicus furinalis; SCSK – south-central skunk; RAC – North-American Raccoon; MexSK-1 – Mexican skunk, variant 1; SE Asia 1, 2 and 3 – diverse dog RABV lineages circulating in the South-East Asia; Africa-2 – dog RABV lineage from the central and western Africa; CASK – California skunk; MexSK-2 – Mexican skunk, variant 2; TXFX – Texas gray fox; COY – coyote; DOG LA – dog RABV from Latin America; AZFX – Arizona gray fox; NCSK – north-central skunk; EUR – fox viruses from moderate latitudes of Eurasia; Aftrca-1 – dog RABV, broadly distributed in Africa; Africa-3 – mongoose RABV from southern Africa; Arctic – Arctic RABV from Eurasia and North America; Arctic-like – Arctic-like RABV from southern and eastern Asia.