Background: eIF2∼P induces translational expression of ATF4, an ISR transcription factor that regulates cell survival during stress.
Results: The LIP isoform of C/EBPβ represses ATF4 transcription during UV stress, reducing ATF4 mRNA available for preferential translation by eIF2∼P.
Conclusion: Transcription of ATF4 is controlled by different stress signaling pathways.
Significance: Differential ISR expression during eIF2∼P is important for alleviation of cell damage during environmental stresses.
Keywords: Gene Expression, Transcription Regulation, RNA Abundance, Translation Control, Transcription Target Genes, Translation Regulation, eIF2 Kinase, Stress Responses, ATF4
Abstract
Different environmental stresses induce the phosphorylation of eIF2 (eIF2∼P), repressing global protein synthesis coincident with preferential translation of ATF4. ATF4 is a transcriptional activator of genes involved in metabolism and nutrient uptake, antioxidation, and regulation of apoptosis. Because ATF4 is a common downstream target that integrates signaling from different eIF2 kinases and their respective stress signals, the eIF2∼P/ATF4 pathway is collectively referred to as the integrated stress response. Although eIF2∼P elicits translational control in response to many different stresses, there are selected stresses, such as exposure to UV irradiation, that do not increase ATF4 expression despite robust eIF2∼P. The rationale for this discordant induction of ATF4 expression and eIF2∼P in response to UV irradiation is that transcription of ATF4 is repressed, and therefore ATF4 mRNA is not available for preferential translation. In this study, we show that C/EBPβ is a transcriptional repressor of ATF4 during UV stress. C/EBPβ binds to critical elements in the ATF4 promoter, resulting in its transcriptional repression. Expression of C/EBPβ increases in response to UV stress, and the liver-enriched inhibitory protein (LIP) isoform of C/EBPβ, but not the liver-enriched activating protein (LAP) version, represses ATF4 transcription. Loss of the liver-enriched inhibitory protein isoform results in increased ATF4 mRNA levels in response to UV irradiation and subsequent recovery of ATF4 translation, leading to enhanced expression of its target genes. Together these results illustrate how eIF2∼P and translational control combined with transcription factors regulated by alternative signaling pathways can direct programs of gene expression that are specifically tailored to each environmental stress.
Introduction
Environmental stresses induce rapid changes in gene expression designed to alleviate cell damage and return cells to homeostasis. For example, protein synthesis is rapidly reduced in response to different stresses, allowing for conservation of cellular resources and a reprogramming of gene expression designed to enhance cell survival. Central to this translational control is a family of protein kinases that phosphorylates the α subunit of the initiation factor eIF2 (eIF2α) (1–4). The eIF2 complexed with GTP and the initiator Met-tRNAiMet associates with the 40 S ribosomal subunit to facilitate recognition of the start codon. Prior to the joining of the 40 and 60 S ribosomal subunits, the GTP associated with eIF2 is hydrolyzed, leading to a release of eIF2-GDP, which is recycled to the active GTP-bound form by a guanine nucleotide exchange factor, eIF2B. Phosphorylation of eIF2α at serine 51 blocks the exchange of eIF2-GDP to eIF2-GTP, resulting in a repression of initiation of protein synthesis (1–4).
Accompanying this repression of global protein synthesis, phosphorylation of eIF2α (eIF2α∼P)2 also leads to preferential translation of selected mRNAs, such as that encoding the transcription factor ATF4 (1, 4–7). In this case, eIF2α∼P delays translation reinitiation, allowing scanning ribosomes to bypass an inhibitory upstream ORF (uORF) located in the 5′-leader of the ATF4 mRNA. Elevated levels of the basic leucine zipper (bZIP) transcription factor ATF4 enhance the expression of target genes involved in amino acid metabolism, nutrient uptake, the redox status of cells, protein processing, and regulation of apoptosis (8–10). Because many different stresses can trigger ATF4 translation through induced eIF2α∼P, this pathway has been referred to as the integrated stress response (ISR) (8). Therefore, the ISR has both translational and transcriptional control features that regulate gene expression designed to alleviate stress damage. These features of the ISR are shared with GCN4 translational control and the general amino acid control pathway in yeast, emphasizing its evolutionarily conserved role in remediation of environmental stresses (11, 12).
In mammals, four different eIF2α kinases can activate the ISR, each responding to a different set of stress conditions. For example, perturbation in protein folding and assembly in the endoplasmic reticulum (ER) induces PERK (EIF2AK3/PEK) phosphorylation of eIF2α (1–4). Nutrient deprivation and UV irradiation stimulate the eIF2α kinase GCN2 (EIF2AK4) (4, 12–15), whereas viral infection and heme deprivation in erythroid cells induce PKR (EIF2AK2) and HRI (EIF2AK1), respectively (16–20). Dysfunctions in these eIF2α kinases are linked with pathologies in multiple organs, emphasizing their important roles in the recognition and alleviation of environmental stress (10, 21–24). For example, loss of PERK leads to Wolcott-Rallison syndrome, which features neonatal diabetes due to loss of islet β-cells, exocrine pancreatic dysfunction, epiphyseal dysplasia, mental retardation, and impairment of hepatic, kidney, and cardiac tissues.
Although a large number of environmental stresses can activate the ISR, there are some reported stress conditions, such as UV irradiation, that do not lead to expression of ATF4 despite robust eIF2α∼P (13, 14, 25). This discordant induction of the ISR was also reported in brain ischemia and non-alcoholic steatohepatitis (26, 27). Along with preferential translation of ATF4, transcriptional induction of ATF4 is a contributor to its increased expression in the ISR (25, 28, 29). Increased ATF4 mRNA helps to replenish the labile transcript for translational expression during eIF2α∼P. However, following stresses, such as UV irradiation, the transcription of ATF4 is repressed, and as a consequence, there are only low levels of ATF4 mRNA available for translation (25). Although eIF2α∼P and the accompanying translational control enhance the resistance of cultured cells to UV treatment, forced expression of ATF4 with the UV insult substantially reduces survival (25). This indicates that differential induction of eIF2α∼P and the ISR in response to certain environmental stresses can be critical for remediation of stress damage. Furthermore, the transcriptional regulation of ATF4 provides an explanation for how induction of eIF2α∼P does not necessarily elicit the same pattern of gene expression for each stress. The levels of each transcript subject to preferential translation by eIF2α∼P can be selectively diminished, thus allowing different sets of mRNAs to be selectively translated depending on the precise stress arrangement (25).
We are interested in the mechanisms controlling the levels of mRNAs encoding for key ISR factors, such as ATF4, and the role that these regulatory processes play in generating unique gene expression profiles directed by eIF2α∼P during different environmental stresses. The ISR is suggested to be regulated by different transcription factors, which can function downstream of alternative stress response pathways. One such bZIP transcription factor whose function can be regulated during stress is C/EBPβ, which facilitates diverse physiological processes, including adipogenesis, immunity, and bone and liver functions (30–32). C/EBPβ can heterodimerize with ATF4, which is suggested to enhance expression of ISR target genes (33–38). Interestingly, translation of the C/EBPβ mRNA can give rise to three different isoforms, namely liver-enriched activating protein (LAP), LAP*, and liver-enriched inhibitory protein (LIP) (30, 39, 40). The carboxyl-terminal bZIP domain is conserved in each of the isoforms, but the LAP/LAP* isoforms contain an amino-terminal trans-activation domain that is missing in the shorter LIP isoform. The expression of these C/EBPβ isoforms is a consequence of the utilization of different start codons in the mRNA, which can be altered during stress (39, 40).
Here we report a new role for C/EBPβ in the regulation of the ISR in response to UV irradiation. We show that C/EBPβ functions as a transcriptional repressor of ATF4 during UV stress. C/EBPβ binds to critical elements in the ATF4 promoter, resulting in its transcriptional repression. As a consequence, there is significantly reduced ATF4 expression accompanying enhanced eIF2α∼P. Furthermore, we show that C/EBPβ expression is enhanced following UV irradiation and that the LIP isoform of C/EBPβ, but not the LAP version, directly represses ATF4 transcription. Loss of the LIP isoform results in increased ATF4 mRNA levels in response to UV irradiation and subsequent recovery of ATF4 translation, leading to enhanced expression of its target genes. Together these results illustrate how a common regulatory process, eIF2α∼P and translational control, combined with different stress-regulated transcriptional mechanisms can direct patterns of gene expression that are specifically tailored to each environmental stress.
EXPERIMENTAL PROCEDURES
Cell Culture and Stress Conditions
Wild-type, CHOP−/−, ATF4−/−, C/EBPβ−/−, and C/EBPβ-ΔuORF mouse embryonic fibroblast (MEF) cells were described previously (40–43). MEF cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1 mm non-essential amino acids, 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C. The ATF4−/− MEF cells were reported to be sensitive to oxidative stress, and therefore culture media for ATF4−/− cells and the wild-type control cells analyzed in parallel in each experiment were supplemented with additional essential amino acids along with 55 μm β-mercaptoethanol (8). MEF cells were grown to about 70% confluence and treated with 40 J/m2 UV-C irradiation (UV Stratalinker 2400) or 1 μm thapsigargin as indicated. In some cases, these stress conditions were combined by first exposing MEF cells to 40 J/m2 UV-C irradiation. Following 1 h of culture, these cells were then treated with 1 μm thapsigargin for up to 6 h as indicated. Measurement of the stability of C/EBPβ mRNA was determined by adding 20 μm actinomycin D 1 h after 40 J/m2 UV-C irradiation, 1 μm thapsigargin, or no stress treatment. Cells were then cultured for an additional 1, 2, or 4 h, and ATF4 and C/EBPβ mRNA levels were measured by quantitative real time PCR (qPCR) as described below.
Preparations of Protein Lysates and Immunoblot Analyses
MEF cells treated with or without stress conditions were washed twice with ice-cold phosphate-buffered saline (pH 7.4) and lysed in a solution containing 50 mm Tris-HCl (pH 7.9), 150 mm NaCl, 0.1% SDS, 100 mm NaF, 17.5 mm glycerol phosphate, and 10% glycerol that was supplemented with protease inhibitors (100 μm phenylmethylsulfonyl fluoride, 0.15 μm aprotinin, 1 μm leupeptin, and 1 μm pepstatin). Lysates were sonicated for 30 s and precleared by centrifugation at 10,000 × g at 4 °C. Protein concentrations were measured using the non-detergent Bio-Rad protein quantification reagent, and equal amounts of the protein preparations were separated by SDS-PAGE using low and high range molecular weight markers (Bio-Rad) for measurements of protein sizes. Separated proteins were then transferred to nitrocellulose filters, which were incubated for 1 h in TBST solution containing 20 mm Tris-HCl (pH 7.9), 150 mm NaCl, 0.2% Tween 20, and 4% (w/v) nonfat dried milk. The filters were incubated overnight with antibody specific to the indicated proteins at 4 °C. Antibody specific for phosphorylated eIF2α at serine 51 was obtained from Cell Signaling Technologies (catalog number 9721), whereas CHOP and β-actin antibodies were obtained from Santa Cruz Biotechnology (catalog number sc-7351) and Sigma (catalog number A5441), respectively. ATF4 antibody was prepared against recombinant protein (44), and the antibody that detected all of the isoforms of C/EBPβ (LAP, LAP*, and LIP) was purchased from Biolegend (clone 1H7). Monoclonal antibody that recognizes total eIF2α (phosphorylated and non-phosphorylated) was provided by Dr. Scott Kimball (Pennsylvania State University College of Medicine, Hershey, PA). The filters were washed three times in the TBST solution followed by incubation with horseradish peroxidase-tagged secondary antibody at room temperature for 1 h. Protein levels were determined with either x-ray film or a LiCOR Odyssey imaging system. Each of the experiments featuring immunoblot analyses was repeated three independent times for validation.
RNA Isolation and Real Time PCR Analyses
MEF cells were treated with the indicated stresses, and total RNA was isolated using TRIzol reagent (Invitrogen). To remove any contaminating DNA, the total RNA preparations were treated with DNase I (Promega) for 30 min at 37 °C. Single strand cDNA synthesis was carried out using the TaqMan reverse transcriptase kit (Applied Biosystems) following the manufacturer's instructions. Levels of mRNA were measured by qPCR by the SYBR Green (Applied Biosystems) method using a Roche Applied Science LightCycler 480 real time PCR system. Levels of target mRNAs were normalized with levels of β-actin transcripts. The primers for measuring ATF4 and C/EBPβ mRNAs were as follows: ATF4: forward primer, 5′-GCCGGTTTAAGTTGTGTGCT-3′; reverse primer, 5′-CTGGATTCGAGGAATGTGCT-3′; C/EBPβ: forward primer, 5′-CGGGTTTCGGGACTTGAT-3′; reverse primer, 5′-GCCCGGCTGACAGTTACAC-3′; and β-actin: forward primer, 5′-TGTTACCAACTGGGACGACA-3′; reverse primer, 5′-GGGGTGTTGAAGGTCTCAAA-3′. The transcript levels expressed from the ATF4 target genes asparagine synthetase (ASNS), cationic amino acid transporter 1 (CAT-1), and CHOP were measured by using qPCR and the following primer sets: ASNS: forward primer, 5′-TTGACCCGCTGTTTGGAATG-3′; reverse primer, 5′-CGCCTTGTGGTTGTAGATTTCAC-3′; CAT-1: forward primer, 5′-CTTTGGATTCTCTGGTGTCCTGTC-3′; reverse primer, 5′-GTTCTTGACTTCTTCCCCTGTGG-3′; and CHOP: forward primer, 5′-CGGAACCTGAGGAGAGAGTG-3′; reverse primer, 5′-CGTTTCCTGGGGATGAGATA-3′. The qPCR measurements were carried out using the LightCycler 480 software to generate Cp values. Values are represented as the mean from three independent experiments with each measurement carried out in triplicate and standard deviations as indicated. Statistical significance was calculated by using the two-tailed Student's t test.
Plasmid Constructions and Luciferase Assays
Transcriptional regulation of ATF4 was measured using a previously reported PATF4-Luc plasmid containing the 2.5-kb mouse ATF4 promoter fused to the firefly luciferase coding region in the pGL3 expression plasmid (25). ATF4 translation control was measured using a PTK-ATF4-Luc plasmid containing the cDNA encoding the 5′-leader of mouse ATF4 mRNA and the ATF4 start codon, which were downstream of a constitutive TK promoter (7).
Deletion analysis of the ATF4 promoter in the PATF4-Luc plasmid was performed by sequentially removing 500-bp segments from the 5′-portion of the ATF4 promoter using divergent 5′-phosphorylated primers to PCR amplify the required construct and subsequent ligation using T4 DNA ligase at room temperature. Additionally, internal deletions of 500-bp segments removing sequences from −2 to −1.5 kb, −1.5 to −1 kb, −1 to −0.5 kb, and −0.5 kb to −1 bp were similarly created in the ATF4 promoter of the PATF4-Luc plasmid. Deletions of predicted C/EBPβ-binding sites involving the indicated sequences in the ATF4 promoter were also constructed in the PATF4-Luc plasmid. Plasmids expressing LAP and LIP isoforms of C/EBPβ were described previously (43).
For the luciferase assays, MEF cells were plated at a density of 105 cells/well in 35-mm plates and grown overnight to a density of 50% cellular confluence. The PATF4-Luc or PTK-ATF4-Luc plasmids were cotransfected with a control Renilla luciferase plasmid at a dilution ratio of 10:1 using FuGENE 6 reagent (Roche Applied Science). 24 h after transfection, the MEF cells were exposed to vehicle alone, 1 μm thapsigargin, or 40 J/m2 UV-C irradiation and then cultured for an additional 12 h. Dual-Luciferase assays were carried out as described by the Promega instruction manual. Luciferase values were derived as the mean of three experiments with the standard deviation indicated. Statistical significance was calculated with the two-tailed Student's t test.
Chromatin Immunoprecipitation
C/EBPβ−/− MEF cells and the wild-type counterpart were plated in 15-cm plates and grown overnight to about 60% confluence. Cells were treated with 40 J/m2 UV-C irradiation followed by 6-h incubation or 1 μm thapsigargin for 6 h or subjected to no stress. Chromatin immunoprecipitations were performed with the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology catalog number 9003) following the manufacturer's protocol. Immunoprecipitation reactions were carried out by using antibodies against C/EBPβ (Biolegend clone 1H7), histone H3 (D2B12) (Cell Signaling Technology catalog number 4620), and rabbit IgG (Cell Signaling Technology catalog number 2729). Immunoprecipitated DNA samples were then analyzed by qPCR. The primers for the segment of DNA analyzed for C/EBPβ binding designated P1, P2, and P3 were as follows: P1: forward primer, 5′-GGGACTGGAGAGTTAGGTTCG-3′; reverse primer, 5′-TGTTTAAGTGACTCACAC-3′; P2: forward primer, 5′-AAGGCTTGAGAGCCAACTGA-3′; reverse primer, 5′-TTCCTCCAGTTCAGCGATTT-3′; and P3: forward primer, 5′-TCGGTTCTGGAAACAACAAA-3′; reverse primer, 5′-GTCACACCTGCCATCTCTTG-3′. Primer sets for RPL30 were provided with the SimpleChIP kit. Values are represented as the mean from three independent experiments with standard deviations as indicated. Statistical significance was calculated by using the two-tailed Student's t test.
RESULTS
ATF4 Expression Is Blocked in Response to UV Irradiation Despite Robust eIF2α∼P
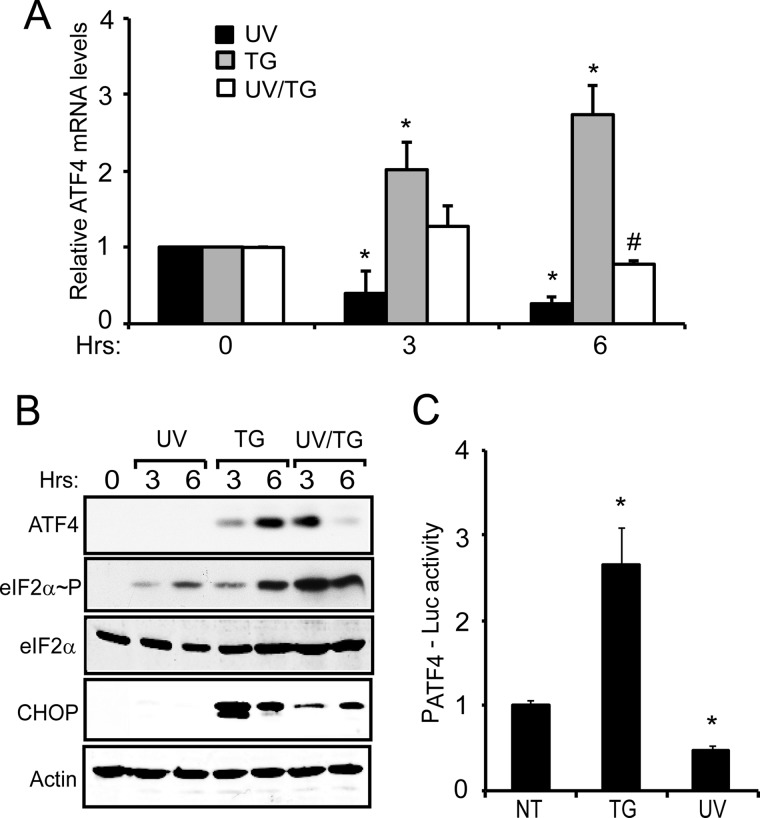
Low ATF4 expression following UV irradiation has been attributed to its transcriptional repression, which diminishes the levels of ATF4 mRNA available for preferential translation during eIF2α∼P (25). These central ideas are illustrated in wild-type MEF cells treated with 40 J/m2 UV-C irradiation or with 1 μm thapsigargin, an inducer of ER stress that enhances both the transcription and translational expression of ATF4 (25). ATF4 mRNA levels as measured by qPCR were lowered about 3-fold 6 h following UV irradiation, whereas ER stress enhanced ATF4 transcript levels by over 2-fold compared with the non-treated cells (Fig. 1A). When UV irradiation was combined with thapsigargin treatment, there was no induction of ATF4 mRNA levels; in fact, following 6 h of this combined stress regimen, there was a significant reduction in ATF4 transcripts. This expression pattern of the ISR indicates that the repressing effects of UV stress are dominant during the progression of the stress response.
FIGURE 1.
Expression of ATF4 is blocked following UV irradiation despite increased eIF2α∼P. A, wild-type MEF cells were treated with either 40 J/m2 UV-C irradiation (UV) or 1 μm thapsigargin (TG) and cultured for 3 or 6 h as indicated. Alternatively, cells were treated with UV-C irradiation for an hour followed by thapsigargin (UV/TG) for the indicated time. 0 h represents no stress treatment. Total mRNA was then isolated from the cells, and the levels of ATF4 mRNA were measured by qPCR. Values obtained are -fold change compared with the no-treatment control. Each experiment was performed three independent times with error bars representing the S.D. * indicates significance with p < 0.05 compared with non-treated control. # indicates a significant difference between the UV and ER stress treatments after 6 h. B, protein lysates were prepared from wild-type MEF cells treated with the conditions as indicated for A. Levels of ATF4, eIF2α∼P, total eIF2α, CHOP, and β-actin were measured by immunoblot analysis using antibodies specific to the indicated proteins. C, the PATF4-Luc reporter plasmid containing 2.5-kb of the ATF4 promoter was transfected into the wild-type MEF cells, which were then treated with 1 μm thapsigargin or 40 J/m2 UV-C irradiation or subjected to no treatment (NT) as indicated. Firefly luciferase activity was measured as described under “Experimental Procedures,” and the luciferase activity relative to the non-treated sample is presented in the histogram along with the S.D. (error bars).
We also measured expression of ATF4 protein by immunoblot analysis in response to UV and ER stress. Both stress treatments increased eIF2α∼P levels with progressive enhancement after 3 and 6 h following the initiation of the stress (Fig. 1B). However, only thapsigargin treatment increased ATF4 protein levels as well as its downstream target gene product, CHOP. Combined treatment of the MEF cells with UV irradiation and thapsigargin led to enhanced eIF2α∼P that exceeded that measured by either stress alone. However, by 6 h of the stress regimen, there was minimal expression of ATF4 protein. Furthermore, the levels of CHOP protein were lowered as compared with thapsigargin treatment alone (Fig. 1B). It is noted that at 3 h of the combined stress treatment there was some expression of ATF4 protein. At this earlier time point, ATF4 mRNA levels were not yet lowered, suggesting some availability of transcript for preferential translation. These protein measurements support the idea that although UV irradiation induces eIF2α∼P this stress condition serves to repress ATF4 expression.
The changes in ATF4 mRNA levels are suggested to be the consequence of transcriptional regulation as wild-type MEF cells containing a luciferase reporter PATF4-Luc, which includes 2.5 kb of the ATF4 promoter, showed over a 2-fold increase in activity during ER stress. By comparison, UV irradiation reduced the PATF4-Luc activity to less than 50% of the non-treated cells (Fig. 1C). Taken together, these results show that ATF4 mRNA is transcriptionally repressed in response to UV irradiation, leading to low levels of ATF4 protein despite enhanced eIF2α∼P.
ATF4 Promoter Contains Elements Responsible for Transcriptional Repression
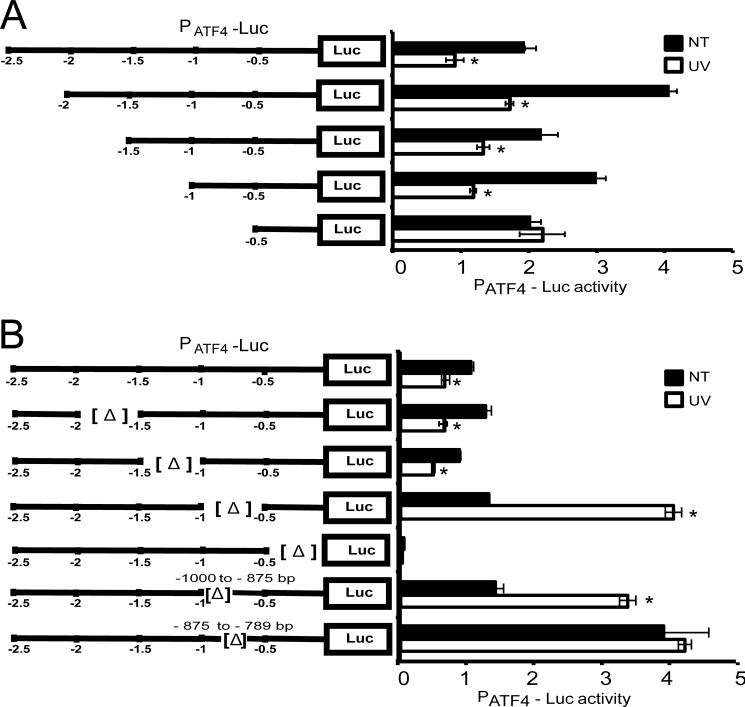
To determine the underlying mechanism for repression of the ATF4 promoter in response to UV irradiation, we processively deleted 500-bp segments from the 5′-end of the 2.5-kb of ATF4 promoter included in the PATF4-Luc plasmid (Fig. 2A). The resulting reporter plasmids were transfected into wild-type MEF cells, which were treated with 40 J/m2 UV irradiation. A 2-fold repression of luciferase activity was seen in cells containing plasmids deleted for the promoter sequences from −2.5 to −1 kb, although there were some significant increases in the PATF4-Luc expression in the non-treated cells with some of the deletion constructs (Fig. 2A). Importantly, deletion of the segment from −1 to −0.5 kb relieved the repression of PATF4-Luc expression with no change in luciferase activity following UV treatment, suggesting that this portion of the ATF4 promoter facilitated transcription repression.
FIGURE 2.
ATF4 promoter contains critical elements for repression in response to UV irradiation. A, the PATF4-Luc reporter plasmid containing 2.5 kb of the ATF4 promoter was transfected into wild-type MEF cells, and following UV irradiation (UV) or no treatment (NT), luciferase activity was measured. In parallel, 0.5-kb segments were sequentially deleted from the 5′-end of the ATF4 promoter in the PATF4-Luc reporter and analyzed for activity in the wild-type cells treated with UV irradiation or subjected to no treatment. PATF4-Luc activity is presented along with the S.D. (error bars). B, internal 0.5-kb deletions were also constructed in the PATF4-Luc reporter and transfected into wild-type MEF cells followed by exposure to UV irradiation or no treatment. Furthermore, smaller deletions were constructed within the −1 to −0.5-kb region of the ATF4 promoter and assayed in wild-type cells in the presence or absence of UV stress. * designates significance (p < 0.05).
In parallel to the processive 5′-deletions of the ATF4 promoter, we also carried out internal 500-bp deletions in the 2.5-kb promoter of the PATF4-Luc reporter (Fig. 2B). Whereas deletions from −2 to −1.5 and −1.5 to −1.0 kb retained repression of the luciferase reporter, removal of a segment from −1 to −0.5 kb changed the UV stress from a repressing signal to one that activates with almost a 4-fold increase in luciferase activity upon UV irradiation (Fig. 2B). As expected, deletion from −0.5 kb to −1 bp removed core promoter sequences, leading to minimal PATF4-Luc expression in both UV-irradiated and non-treated cells.
The ATF4 promoter sequences from −1.0 to −0.5 kb are suggested to be central for repression of ATF4 in response to UV irradiation. Interestingly, included within this promoter segment are two predicted C/EBPβ-binding elements situated at −950 to −935 bp (TAAATAGCAATCAAT) and at −874 to −859 bp (TTGCAAATAATCACT) that reside in this portion of the ATF4 promoter. We constructed smaller internal deletions that removed one or the other of the predicted C/EBPβ-binding sequences. A deletion from −1000 to −875 bp led to over a 2-fold increase in PATF4-Luc expression in response to UV irradiation, which recapitulated the key findings from the larger −1.0 to −0.5-kb internal deletion (Fig. 2B). Deletion of the −875 to −789-bp region triggered high basal luciferase expression, which was retained even with UV stress. These results suggest that there are key regulatory elements in the ATF4 promoter, from approximately −1000 to −789 bp, that facilitate repression in response to UV irradiation.
C/EBPβ Represses ATF4 Promoter
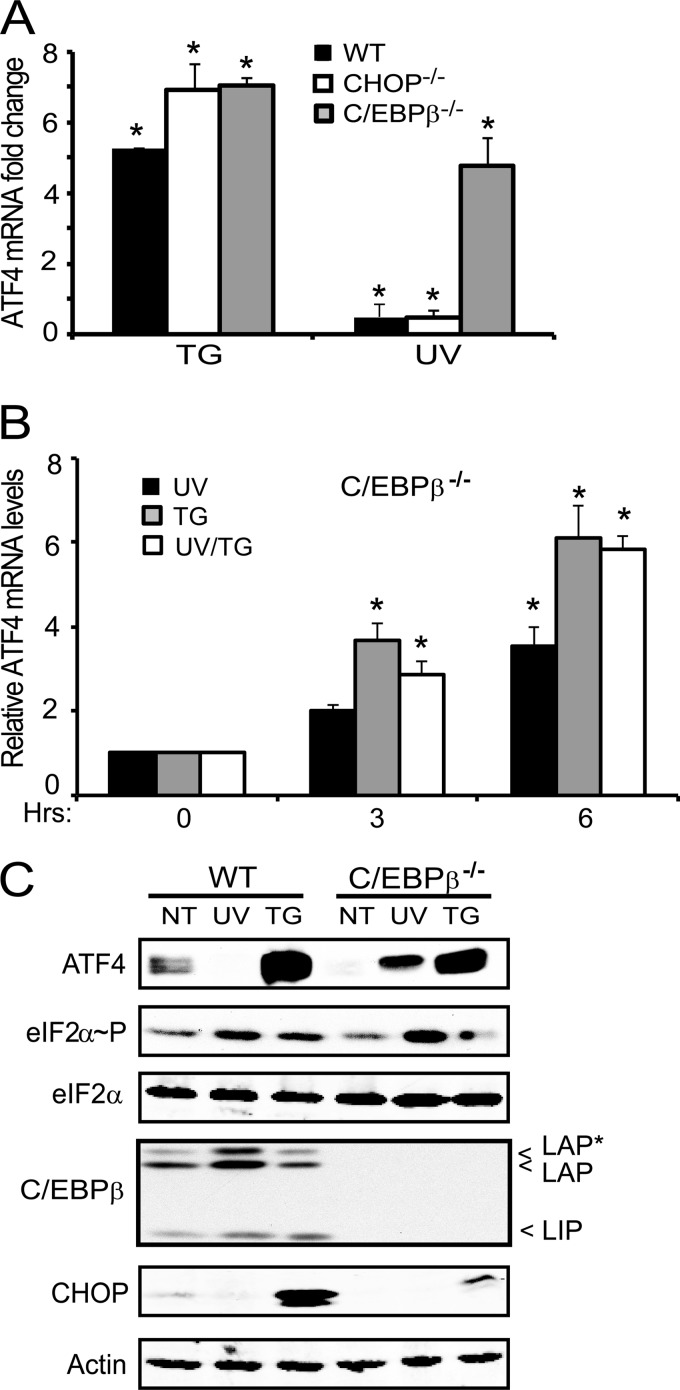
Our promoter deletion analyses suggested that C/EBPβ contributes to transcriptional repression of ATF4. To test this idea, we treated C/EBPβ−/− MEF cells and the wild-type counterpart with UV irradiation or thapsigargin and measured the ATF4 transcript and protein levels. The C/EBPβ−/− MEF cells displayed a 4-fold increase in ATF4 mRNA levels 6 h after the UV treatment compared with non-treated cells (Fig. 3A). By comparison, wild-type and CHOP−/− MEF cells showed reduced amounts of ATF4 transcript in response to UV irradiation. Each of these cell lines had increased ATF4 mRNA levels in response to ER stress; in fact, the C/EBPβ−/− cells showed almost a 7-fold enhancement of ATF4 mRNA upon thapsigargin treatment that was modestly, albeit significantly, higher than that in wild-type cells (Fig. 3A). Previously, we noted that the repressing effects of UV irradiation were dominant in combination stress treatments in wild-type cells (Fig. 1, A and B). However, in C/EBPβ−/− cells, the combination of UV and thapsigargin treatments increased ATF4 mRNA to levels similar to levels measured for ER stress alone (Fig. 3B).
FIGURE 3.
C/EBPβ is required for reduced ATF4 mRNA in response to UV irradiation. Wild-type, CHOP−/−, and C/EBPβ−/− MEF cells were treated with 1 μm thapsigargin (TG) or with 40 J/m2 UV-C irradiation (UV) and cultured for 6 h. A, the levels of ATF4 mRNA were measured by qPCR, and the -fold change in the transcript levels is represented relative to cells not treated with stress with the S.D. indicated by error bars. B, C/EBPβ−/− MEF cells were treated with UV irradiation, thapsigargin, or both (UV/TG) and cultured for 3 or 6 h. Values are relative to the no-treatment control (0), and the S.D. is indicated by error bars. C, protein lysates were prepared from wild-type and C/EBPβ−/− MEF cells subjected to UV irradiation, thapsigargin, or no treatment (NT), and the indicated protein levels were measured by immunoblot analysis. The LAP and LIP isoforms of C/EBPβ are indicated to the right of the panel. Results are representative of three independent experiments. * indicates significance with p < 0.05.
Immunoblot analyses of ATF4 protein from lysates prepared from C/EBPβ−/− cells treated with UV irradiation also showed increased ATF4 levels, whereas ATF4 protein was absent in the similarly stressed wild-type cells (Fig. 3C). During ER stress, increased ATF4 protein was observed in both wild-type and C/EBPβ−/− cells. As expected, both LIP and LAP forms of C/EBPβ were not present in the C/EBPβ-deleted cells. Levels of CHOP protein were significantly diminished in the C/EBPβ−/− cells during ER stress compared with wild type and absent in the mutant cells subjected to UV irradiation (Fig. 3C). Results of this experiment are consistent with our measurements of ATF4 mRNA, indicating that C/EBPβ serves to repress ATF4 expression.
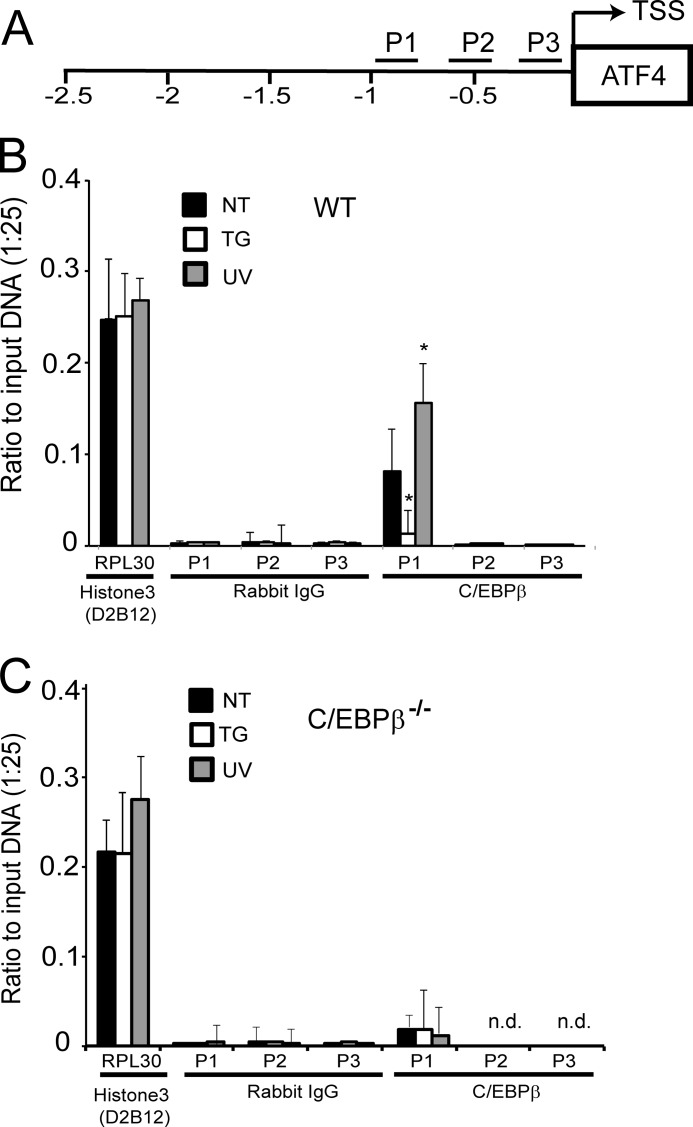
To determine whether C/EBPβ directly binds in vivo to the regulatory elements in the ATF4 promoter, we carried out ChIP analysis using antibody that specifically recognizes C/EBPβ. We considered three sites in the ATF4 promoter: P1 that includes the two predicted C/EBPβ-binding sites (−978 to −800 bp) along with two flanking regions designated P2 (−628 to −470 bp) and P3 (−334 to −194 bp) (Fig. 4A). There was specific C/EBPβ binding at the P1 segment that was absent using control antibody (Fig. 4B). The C/EBPβ association at the P1 portion of the ATF4 promoter was enhanced in MEF cells following UV irradiation when ATF4 mRNA levels are low, and there was decreased binding of C/EBPβ to the P1 segment during ER stress when ATF4 transcript levels are elevated. As expected, no significant C/EBPβ binding to the P1 segment of the ATF4 promoter was found in the C/EBPβ-deleted cells (Fig. 4C). Additionally, C/EBPβ was not associated with the P2 or P3 portion of the ATF4 promoter in the ChIP experiments using either C/EBPβ-specific or control antibody. As an additional control, histone H3 Lys-4-specific antibody was observed to bind to a portion of the RPL30 gene in either wild-type or C/EBPβ−/− cells independently of stress arrangements (Fig. 4, B and C). These results support the model that C/EBPβ binds to a specific segment of the ATF4 promoter following UV stress, triggering repression of transcription. In conditions such as ER stress where ATF4 transcription is activated, there is minimal binding of C/EBPβ at this ATF4 promoter region.
FIGURE 4.
C/EBPβ binds to specific elements in ATF4 promoter. A, schematic representation of the regions of ATF4 promoter analyzed for C/EBPβ binding by ChIP. TSS indicates the transcriptional start site, and the promoter region is illustrated from −2.5 kb to the +1 position. The region designated P1 includes the sequence −978 to −800 bp in the ATF4 promoter, and P2 and P3 represent regions −628 to −470 bp and −334 to −194 bp, respectively. B, 6 h after exposure to 40 J/m2 UV-C irradiation (UV) or 1 μm thapsigargin (TG) or no treatment (NT), wild-type (WT; B) and C/EBPβ−/− (C) cells were analyzed by ChIP analyses for C/EBPβ binding to the P1, P2, and P3 regions of the ATF4 promoter. The immunoprecipitated DNA was analyzed by qPCR using primer sets specific for each promoter region. C/EBPβ indicates that antibody specific to this transcription factor was used in the ChIP assay; rabbit IgG antibody was used as a control. ChIP analyses were also carried out with the control histone H3 Lys-4 (D2B12) antibody and were analyzed by qPCR using primer sets for RPL30. Data are represented as a ratio of the input sample (1:25) and are the mean and S.D. (error bars) of three different experiments. * indicates significance with p < 0.05. n.d. indicates that the C/EBPβ binding to the P2 and P3 was not detected in the C/EBPβ−/− cells.
Expression of C/EBPβ Isoforms Is Differentially Regulated in Response to UV and ER Stress
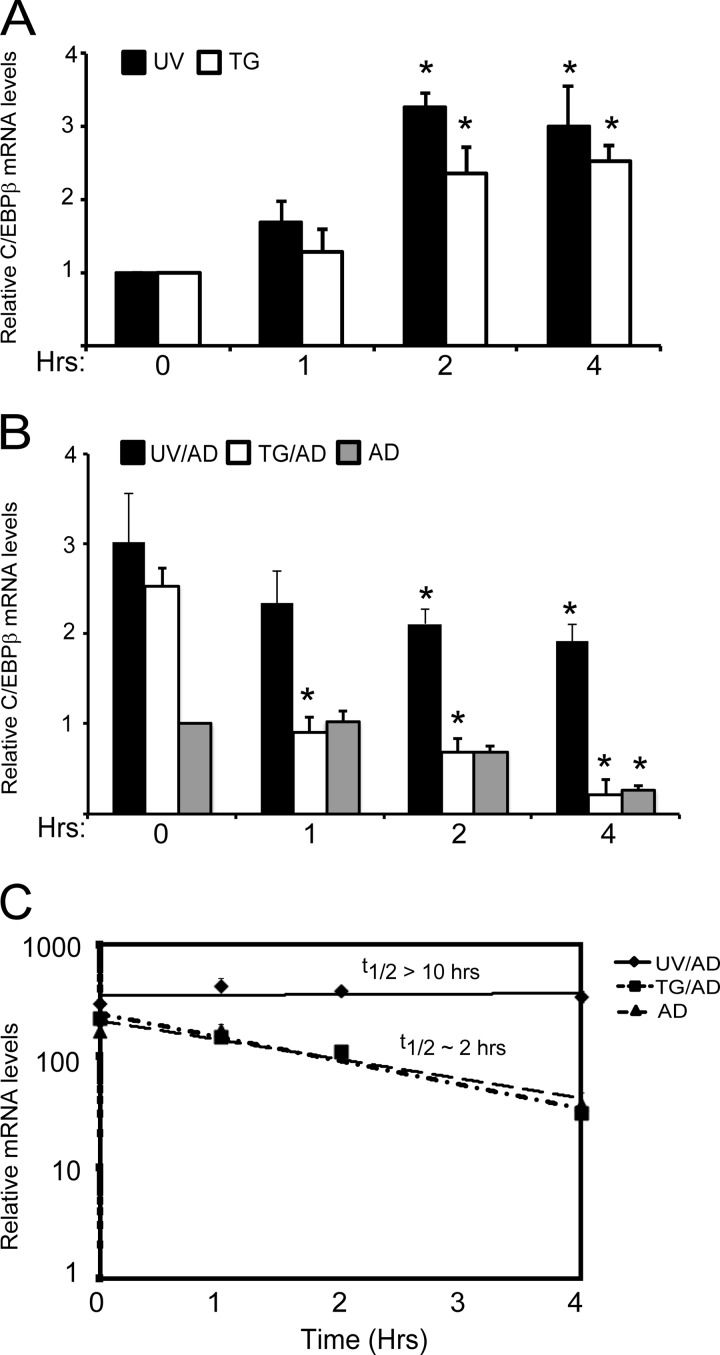
Measurements of C/EBPβ mRNA levels in response to either UV or ER stress showed increases in transcript levels 2 h after the initiation of the stress with UV irradiation eliciting the largest enhancement in C/EBPβ expression (Fig. 5A). Interestingly, measurements of the turnover of C/EBPβ mRNA by treatment of the wild-type MEF cells with actinomycin D, a potent inhibitor of RNA polymerase II, prior to the stress exposure indicated that UV exposure increased the half-life of the C/EBPβ transcripts to greater than 10 h (Fig. 5, B and C). By comparison, the half-life of C/EBPβ mRNA was ∼2 h in the cells treated with thapsigargin or subjected to no stress. Together these studies suggest that UV and ER stress can differentially affect the expression of C/EBPβ.
FIGURE 5.
C/EBPβ mRNA is stabilized following UV irradiation. A, wild-type MEF cells were exposed to 1 μm thapsigargin (TG) or 40 J/m2 UV-C irradiation (UV) and cultured for up to 4 h as indicated. RNA was prepared from these cells, and C/EBPβ mRNA levels were measured by qPCR. The amount of C/EBPβ transcript is presented relative to the no-treatment control (0), and the S.D. is indicated by error bars. * indicates significance with p < 0.05. B, measurements of C/EBPβ mRNA half-life were carried out by first treating cells with UV irradiation or thapsigargin. 1 h after the initiation of the stress regimen, transcription was blocked by treating the cells with 20 μm actinomycin D (UV/AD or TG/AD), and then cells were cultured for up to an additional 4 h. Alternatively, cells were treated with actinomycin D (AD) alone. C/EBPβ mRNA levels were measured by qPCR at the indicated time intervals. Values are representative of the mean with the S.D. indicated by error bars. C, the half-life of the C/EBPβ mRNA for each of the stress arrangements was determined by plotting the transcript levels versus the length of time of the actinomycin D treatment in a semilogarithmic graph.
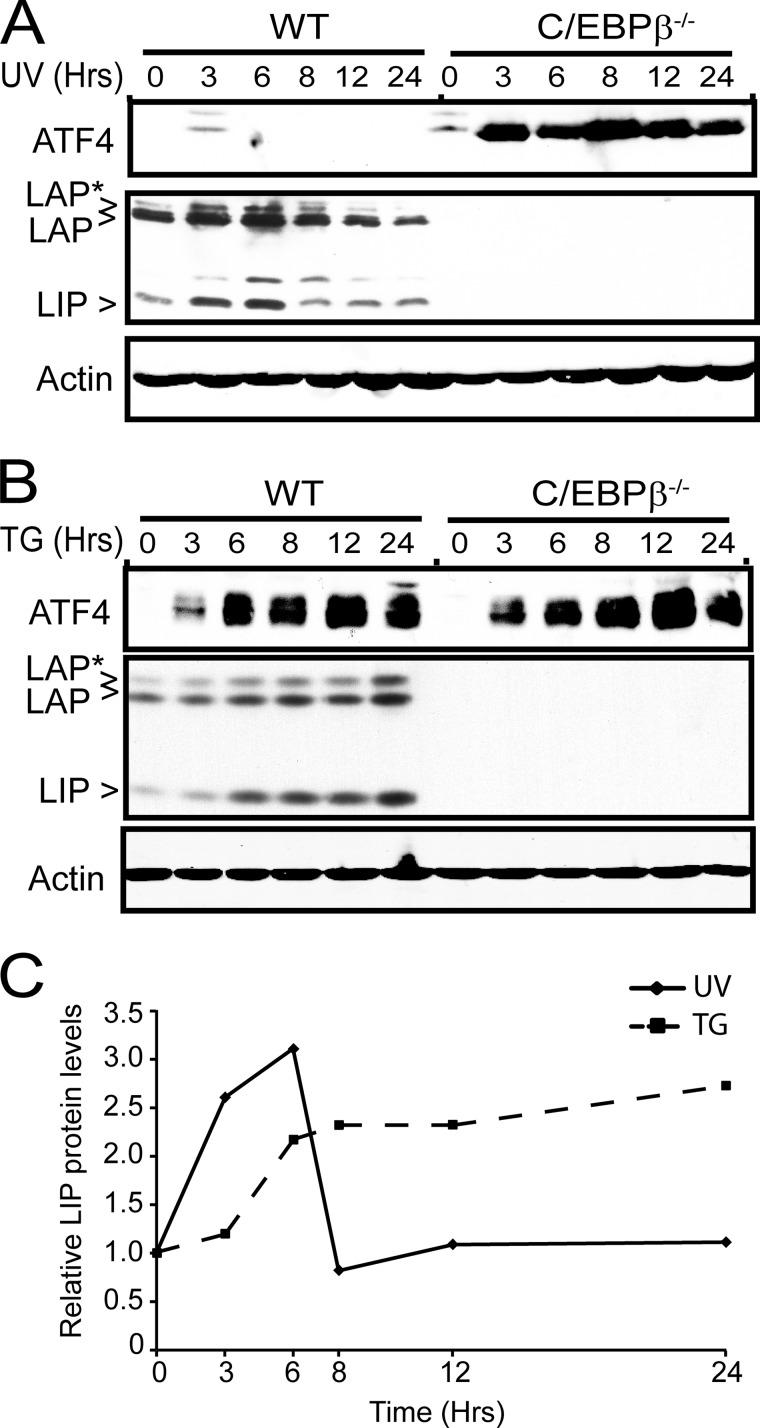
We next measured C/EBPβ proteins, which can exist as three isoforms: LAP and LAP*, which include an extended amino terminus, and LIP, which is devoid of this transcriptional activation region and has been observed to inhibit transcription (30–32, 45). To determine whether any significant change occurs in the expression of the C/EBPβ isoforms following thapsigargin treatment or UV irradiation, we treated wild-type and C/EBPβ−/− MEF cells with these stress arrangements and measured the changes in the levels of this transcription factor for up to 24 h. In response to UV irradiation, there was a significant increase in LIP levels after 3 and 6 h of UV treatment, returning to non-stressed amounts after 8 h (Fig. 6A). ER stress resulted in no change in LIP expression after 3 h of thapsigargin exposure, but by 6 h, there was an increase in LIP protein levels that extended for up to 24 h of stress treatment (Fig. 6B). These results indicate that expression of LIP, a known transcriptional inhibitor, can be differentially regulated during UV and ER stress (Fig. 6C). Following 3 h of UV irradiation, LIP levels are sharply enhanced, which coincides with repression of ATF4 transcription. By comparison, induction of LIP expression does not occur until later in the ER stress regimen, 6 h after the onset of thapsigargin treatment.
FIGURE 6.
LIP isoform of C/EBPβ is differentially expressed during UV and ER stress. CEBPβ−/− MEF cells and the wild-type counterpart were treated with 40 J/m2 UV-C irradiation (UV) (A) or 1 μm thapsigargin (TG) (B) and were cultured for up to 24 h as indicated. Protein lysates were prepared from the treated cells, and the levels of ATF4, LIP, LAP, and β-actin were measured by immunoblot analyses. Each panel is representative of three independent experiments. C, the levels of the LIP isoform of C/EBPβ were quantified by densitometry and are represented as relative levels of the LIP band as compared with the no-treatment control (0).
LIP Is a Potent Repressor of ATF4 Transcription
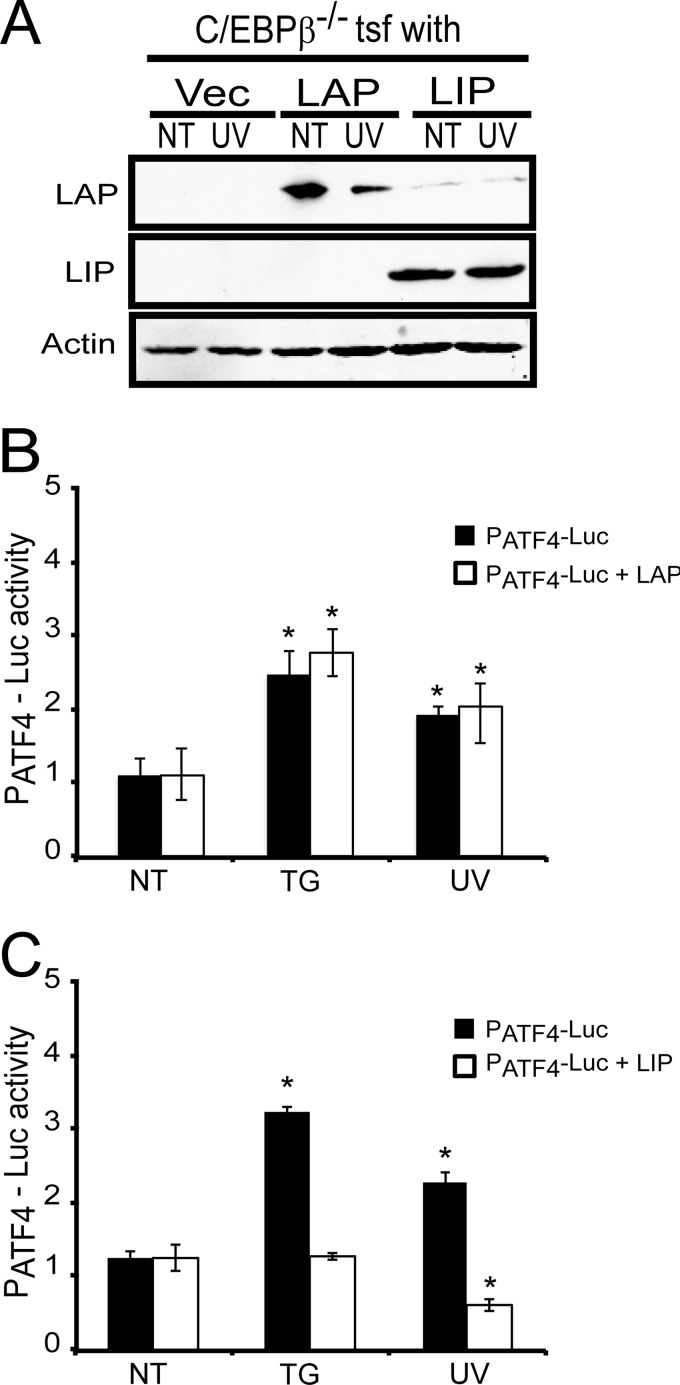
Our studies suggest that the LIP isoform of C/EBPβ can function as a repressor of ATF4 transcription. We addressed this idea by two experimental approaches. First, we transfected plasmids specifically expressing either the LIP or LAP isoforms into the C/EBPβ−/− MEF cells and measured the activity of the ATF4 promoter using the PATF4-Luc reporter. In cells expressing only LAP (Fig. 7A), there was about a 2-fold increase in PATF4-Luc activity following UV irradiation (Fig. 7B). This level of induced luciferase activity was similar to that measured in the C/EBPβ−/− cells transfected with the expression vector alone. The levels of PATF4-Luc activity were also increased over 2-fold upon thapsigargin treatment independently of LAP expression (Fig. 7B). By comparison, expression of LIP in the C/EBPβ−/− cells restored repression of the ATF4 promoter with a 50% reduction in luciferase activity with UV irradiation (Fig. 7, A and C). Interestingly, expression of LIP in the C/EBPβ−/− cells also blocked the activation of the ATF4 promoter in response to ER stress (Fig. 7C), suggesting that low LIP expression in the early phase of ER stress is critical for induction of ATF4 mRNA (Fig. 6B).
FIGURE 7.
LIP is a potent repressor of ATF4 transcription. A, C/EBPβ−/− MEF cells were cotransfected with the PATF4-Luc reporter and plasmids specifically expressing either the LIP or LAP isoforms of C/EBPβ or the parent vector (Vec). The transfected cells were treated with UV irradiation (UV) or subjected to no treatment (NT), and the levels of LAP, LIP, and β-actin were measured by immunoblot analyses. Levels of PATF4-Luc activity were also measured in the cells expressing LAP (PATF4-Luc + LAP) (B) or LIP (PATF4-Luc + LIP) (C) as compared with the cells containing PATF4-Luc and the expression vector alone (PATF4-Luc). Luciferase activity is presented in the histograms with the luciferase activity in the non-treated wild-type cells being represented as a value of 1. Values were derived from three independent experiments with the S.D. indicated by error bars. * indicates significance with p < 0.05. Tsf, transfection of cells with the indicated expression plasmids.
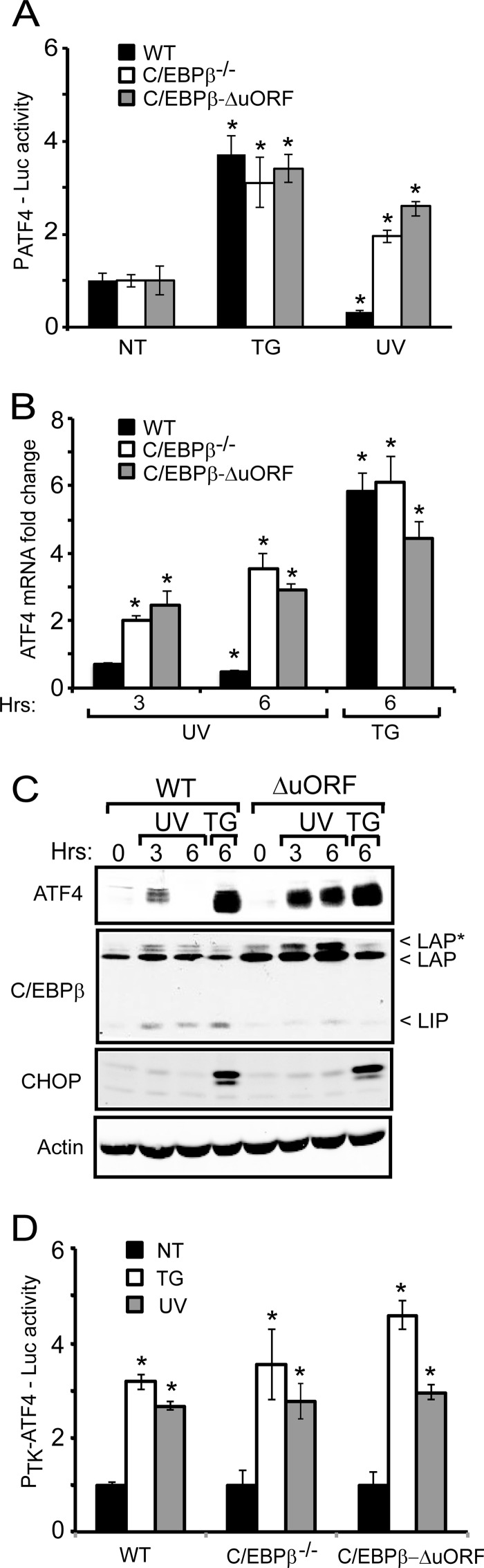
Our second experimental approach to address the role of LIP in the repression of ATF4 transcription centered on previous reports that a short uORF in the C/EBPβ mRNA is essential for expression of the LIP isoform (39, 40). This uORF can inhibit translation initiation of LAP and instead enhance ribosomal scanning and subsequent reinitiation at the downstream LIP. As a consequence, MEF cells containing a mutation in the initiation codon of the uORF (C/EBPβ-ΔuORF) lose the ability to express LIP, resulting in only LAP and LAP* being present in the cells (40). We transfected the PATF4-Luc reporter into the C/EBPβ−/− and C/EBPβ-ΔuORF MEF cells and the wild-type counterpart and measured the ATF4 promoter activity in response to either treatment with UV irradiation or thapsigargin. In response to the UV stress, luciferase activity was increased by over 2-fold in C/EBPβ−/− and C/EBPβ-ΔuORF cells, whereas PATF4-Luc expression was sharply decreased in the wild-type cells (Fig. 8A). There was no difference in the activity of the ATF4 promoter in the mutant and wild-type MEF cells during ER stress. The levels of ATF4 mRNA in the C/EBPβ-ΔuORF MEF cells were significantly enhanced in response to UV irradiation, which was similar to the levels found in the C/EBPβ−/− cells (Fig. 8B). By comparison, the wild-type cells showed a sharp reduction in ATF4 transcripts following UV stress. During ER stress, the C/EBPβ−/−, C/EBPβ-ΔuORF, and wild-type cells each showed over a 4-fold increase in ATF4 mRNA levels.
FIGURE 8.
Loss of LIP in C/EBPβ-ΔuORF cells alleviates repression of ATF4 transcription. A, wild-type, C/EBPβ−/−, and C/EBPβ-ΔuORF MEF cells were transfected with the PATF4-Luc plasmid and treated with either with 1 μm thapsigargin (TG) or 40 J/m2 UV-C irradiation (UV) or subjected to no stress treatment (NT). PATF4-Luc expression was measured and is represented in the histogram with the non-treated cells indicated as a value of 1. Values were determined from three independent experiments with the S.D. indicated by error bars. B, the wild-type (WT), C/EBPβ−/−, and C/EBPβ-ΔuORF cells were treated with UV or thapsigargin stress for up to 6 h, and the levels of ATF4 mRNA were determined by qPCR. Mean values are presented in the histograms with the S.D. indicated by error bars. C, alternatively, the levels of the indicated proteins in the stressed wild-type (WT) and C/EBPβ-ΔuORF (ΔuORF) cells were measured by immunoblot analyses. The zero time indicates no stress treatment. D, levels of ATF4 translational control were measured in wild-type, C/EBPβ−/−, and C/EBPβ-ΔuORF cells that were transfected with the PTK-ATF4-Luc reporter. Following UV or thapsigargin treatment, luciferase activity was measured and is presented in the histograms relative to no stress treatment (NT) with a value of 1. The luciferase measurements were from three independent experiments with the S.D. indicated by error bars.
We also measured ATF4 protein in wild-type and C/EBPβ-ΔuORF cells and found that there was an increase in ATF4 protein following 3 and 6 h of UV irradiation (Fig. 8C). In the matched wild-type MEF cells, we observed only low levels of ATF4 expression 3 h after the UV treatment and no ATF4 protein 6 h after initiating the stress. As expected, LIP was not detected in the C/EBPβ-ΔuORF cells. Furthermore, CHOP protein was significantly diminished in C/EBPβ-ΔuORF cells in response to UV irradiation, whereas during ER stress, there were similar levels of induced CHOP in both the wild-type and C/EBPβ-ΔuORF cells. This finding suggests that LIP, but not LAP, can facilitate CHOP function.
Finally, we measured preferential translation of ATF4 in the wild-type, C/EBPβ−/−, and C/EBPβ-ΔuORF cells in which eIF2α∼P was induced by either UV or ER stress (Fig. 8D). The ATF4 translational control was determined by transfecting into these cells a previously described plasmid encoding the 5′-leader of the ATF4 mRNA that includes the uORFs between the constitutive TK promoter and the firefly luciferase reporter (7). There was a significant increase in the luciferase activity in each cell line in response to either UV irradiation or thapsigargin (Fig. 8D). This indicates that C/EBPβ is not required for preferential translation of ATF4 in response to eIF2α∼P, and if the ATF4 mRNA is available following UV irradiation, there will be high levels of synthesized ATF4 protein. Taken together these experiments demonstrate that LIP is critical for repression of ATF4 transcription in response to UV irradiation. Furthermore, LAP is not required for activation of the ATF4 promoter in response to ER stress.
Loss of C/EBPβ Isoform LIP Increases Expression of ATF4 Target Genes
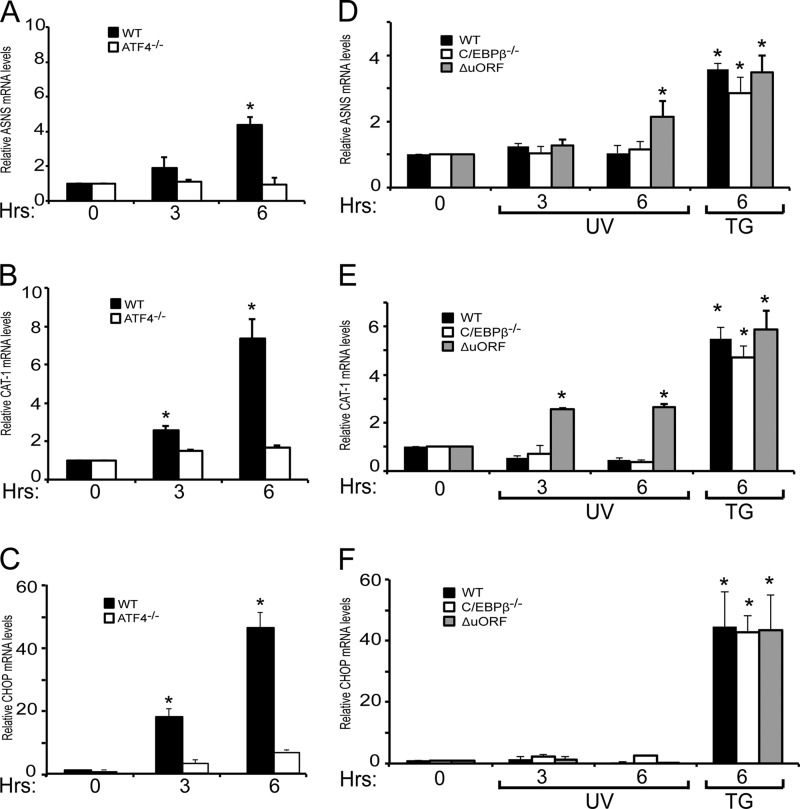
The absence of LIP led to increased ATF4 mRNA and protein levels in both C/EBPβ−/− and C/EBPβ-ΔuORF MEF cells in response to UV irradiation. We next wanted to determine whether expression of key ATF4 target genes in the ISR also increased with the elevated ATF4 expression in the LIP-deficient cells. We measured three well characterized ISR genes: ASNS, which catalyzes the conversion of aspartate to asparagine; CAT-1, an amino acid transporter, and CHOP, which encodes a bZIP transcription factor that can facilitate apoptosis (Fig. 9) (9, 10, 33, 46). As illustrated in Fig. 9, A, B, and C, ATF4 was required for full induction of each of these mRNAs in response to ER stress. Next we measured ASNS, CAT-1, and CHOP mRNAs in the wild-type, C/EBPβ−/−, and C/EBPβ-ΔuORF MEF cells treated with UV irradiation or thapsigargin. The levels of each of these three transcripts were not increased in either wild-type or C/EBPβ−/− cells when treated with UV irradiation, whereas ASNS and CAT-1 mRNAs increased significantly by 6 h in the C/EBPβ-ΔuORF cells (Fig. 9, D and E). Transcription of these genes was uniformly increased among these MEF cells in response to ER stress. These results suggest that increased ATF4 protein levels lead to increased expression of two of its key target genes. The fact that these genes were induced in C/EBPβ-ΔuORF cells but not in the C/EBPβ−/− cells is consistent with reports that LAP heterodimerizes with ATF4 and facilitates transcription of ISR promoters, such as ASNS (33, 34). The levels of CHOP mRNA did not significantly increase in the C/EBPβ-ΔuORF cells in response to UV irradiation (Fig. 9F). As will be highlighted further in the “Discussion,” this observation is consistent with previous reports that, like ATF4, prior treatment with UV irradiation blocks induced CHOP transcription by other stress treatments that enhance eIF2∼P and ATF4 expression (47, 48).
FIGURE 9.
Alleviation of ATF4 repression in C/EBPβ-ΔuORF cells causes increased expression of ATF4 target genes in response to UV irradiation. Wild-type and ATF4−/− MEF cells were treated with 1 μm thapsigargin for up to 6 h, and the levels of ASNS (A), CAT-1 (B), and CHOP (C) mRNAs were measured by qPCR. Wild-type, C/EBPβ−/−, and C/EBPβ-ΔuORF cells were treated with 40 J/m2 UV-C irradiation (UV) or thapsigargin (TG) and cultured for up to 6 h as indicated. The levels of ASNS (D), CAT-1 (E), and CHOP (F) mRNAs were measured by qPCR. Values are presented relative to the no-treatment controls (0), and the S.D. for each is indicated by an error bar. * indicates significance with p < 0.05.
DISCUSSION
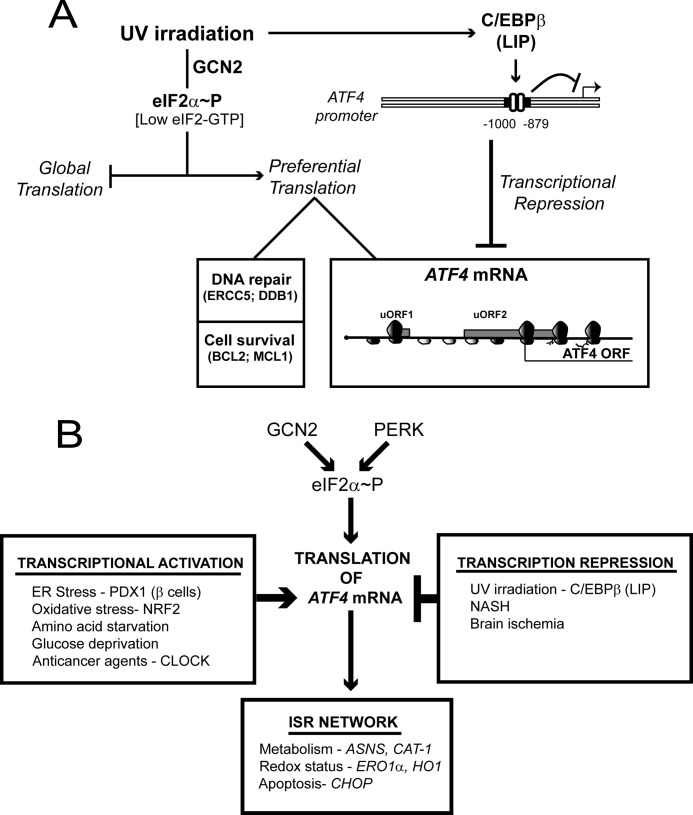
This study provides mechanistic insight into how ATF4 expression is repressed in response to UV irradiation despite induction of eIF2α∼P. As highlighted in the model in Fig. 10A, transcription of ATF4 is repressed following UV irradiation, and therefore, there are low levels of ATF4 mRNA available for translation during eIF2α∼P (Fig. 1) (25). This differs from environmental stresses that increase ATF4 synthesis, such as those afflicting the ER where there is activation of ATF4 transcription, thus further enhancing the levels of ATF4 mRNA for preferential translation by eIF2α∼P. Central to the repression of ATF4 transcription is the LIP isoform of C/EBPβ (Figs. 4, 7, 8, and 10A). The ATF4 promoter contains elements that bind C/EBPβ, and this association is enhanced following UV irradiation (Figs. 4 and 10A). Sequential 5′-truncations as well as internal deletions of the ATF4 promoter indicate that sequences situated between −1000 and −789 bp facilitate repression of ATF4 transcription in response to the UV stress (Fig. 2). Within this repressing region of the ATF4 promoter are binding sites for C/EBPβ, which encodes three isoforms of the bZIP transcriptional regulator, LAP, LAP*, and LIP, produced by differential selection of initiation codons during translation (39, 40). Deletion of C/EBPβ also negated the inhibition of ATF4 transcription following UV irradiation, turning this stress condition instead into an inducer of ATF4 mRNA and protein expression (Fig. 3).
FIGURE 10.
LIP repression of ATF4 transcription reduces levels of ATF4 mRNA available for preferential translation in response to eIF2α∼P during UV stress. A, model for LIP repression of ATF4 expression during UV stress. In response to UV irradiation, GCN2 phosphorylation of eIF2α lowers the levels of eIF2-GTP, resulting in reduced global translation. Additionally, eIF2α∼P leads to preferential translation of genes involved in repair of damaged DNA and those that thwart apoptosis, although the underlying mechanisms have not yet been determined (61). UV irradiation triggers repressed transcription of the ATF4 gene by increased C/EBPβ association at the ATF4 promoter sequences between −1000 and −879 bp. The LIP isoform of C/EBPβ is central for ATF4 repression, and this regulation is suggested to involve LIP engagement with promoter target sequences when LIP is dimerized with other bZIP proteins. The mechanism by which UV irradiation triggers increased LIP regulation of the ATF4 promoter is suggested to involve enhanced stabilization of C/EBPβ mRNA. The resulting lowered ATF4 mRNA levels diminish the amount of transcripts available for preferential translation in response to eIF2α∼P. The resulting loss of ATF4 expression during UV stress impedes the induction of its ISR target genes. B, a combination of transcriptional and translational control of ATF4 directs the gene expression program of the ISR. The eIF2 kinase GCN2 is activated by nutritional deprivation or UV irradiation, whereas PERK is regulated by ER stress. The resulting induced eIF2α∼P can lead to preferential translation of ATF4 by a mechanism involving delayed ribosome reinitiation, which ribosomes to bypass an inhibitory uORF in the ATF4 mRNA. Activation of ATF4 transcription by many different stresses enhances the amount of ATF4 mRNA available for translation in response to eIF2α∼P. Transcription factors that activate the ATF4 promoter include PDX1 in islet β-cells of the pancreas upon ER stress, NRF2 in response to oxidative stress, and CLOCK, which facilitates resistance to anticancer agents cisplatin and etoposide. As a consequence, there will be enhanced levels of ATF4 that directly activate the transcription of ISR target genes involved in metabolism, the redox status of cells, and regulation of apoptosis. Examples of target genes for each ISR category are illustrated. Alternatively, the ATF4 promoter can be repressed by a different set of stress conditions. The LIP isoform of C/EBPβ directly facilitates repression of ATF4 transcription in response to UV irradiation. This would result in low levels of ATF4 mRNA available for preferential translation during UV stress despite high levels of eIF2α∼P, thus lowering the expression of the ATF4 target genes in the ISR. NASH, non-alcoholic steatohepatitis.
The shorter version of C/EBPβ, LIP, contains a bZIP domain that is important for DNA binding but is missing the amino-terminal activation domain. Hence, LIP is a documented repressor of transcription (30–32, 45). Our ChIP analysis of the ATF4 promoter indicates that C/EBPβ binds directly within the repressing region with enhanced association following UV treatment (Fig. 4). Although the ChIP experiments did not distinguish whether the LIP and LAP versions of C/EBPβ associated with the ATF4 promoter, two lines of evidence support the idea that LIP represses the transcription of ATF4 (Fig. 10A). First, expression of LIP, but not LAP, restored repression of ATF4 transcription in C/EBPβ−/− cells subjected to UV irradiation (Fig. 7). Second, ATF4 transcription was not repressed upon UV irradiation in C/EBPβ-ΔuORF cells, which express the LAP and LAP* isoforms but not LIP (Fig. 8). In fact, UV stress in the C/EBPβ-ΔuORF cells led to a significant increase of ATF4 mRNA and protein levels in response to UV irradiation.
A consequence of reduced ATF4 expression during UV stress is the loss of activation of its target genes despite robust eIF2α∼P and the accompanying repression of global translation (Fig. 10A) (25). However, increased ATF4 expression was observed in the C/EBPβ-ΔuORF cells subjected to UV irradiation, which resulted in enhanced transcription of ISR target genes ASNS and CAT-1 (Fig. 9, A–C). Therefore, UV irradiation can trigger significant activation of the ISR target genes when ATF4 expression is restored upon eIF2α∼P.
It is noteworthy that there was no increase in the ATF4 target gene CHOP in C/EBPβ-ΔuORF cells following UV stress. Previously, Schmitt-Ney and Habener (47) reported that UV irradiation is a potent repressor of CHOP expression, and like ATF4, prior treatment with UV irradiation blocks induced CHOP transcription by other stress treatments, such as ER stress and nutrient deprivation. Central to this repression is the first exon of CHOP as inclusion of this region of the CHOP gene into a reporter containing the CHOP promoter represses transcription in response to UV irradiation (47). This would explain the absence of induced CHOP transcription in the C/EBPβ-ΔuORF cells where ATF4 is activated by UV stress; i.e. ATF4 activation of CHOP transcription is blocked by repressing factors that can function via an exon region of CHOP. Furthermore, this suggests that although LIP is a potent repressor of ATF4 in response to UV irradiation this isoform of C/EBPβ does not directly contribute to repression of CHOP. It was also reported that C/EBPβ can dimerize with CHOP, which allows increased stability and nuclear targeting of CHOP (43, 49). Therefore, C/EBPβ can regulate multiple steps in the ISR, including both regulation of ATF4 transcription and the function of its downstream effector, CHOP.
Transcriptional Regulation Combined with Translational Control of ATF4 Allows for Different Patterns of ISR Gene Expression
Transcriptional regulation of ATF4 provides a new dimension to the ISR. The half-lives of ATF4 mRNA and protein are short (25, 50); therefore, the activity of ATF4 is tightly linked to its synthesis, namely the transcription of ATF4 and its translation with the latter being dictated by the status of eIF2α∼P. Activation of ATF4 transcription leads to more mRNA available for preferential translation during eIF2α∼P (Fig. 10B). Alternatively, repression of ATF4 leads to lower mRNA levels, thus diminishing synthesis of ATF4. In this way, eIF2∼P can repress global protein synthesis, allowing for conservation of resources, but ATF4 expression and the ISR can be differentially expressed depending on the nature of the stress condition. In addition to UV irradiation for which we showed that LIP represses ATF4 transcription, brain ischemia and Non-alcoholic steatohepatitis were reported to induce eIF2α∼P yet trigger low ATF4 expression (Fig. 10B) (26, 27). Therefore, ATF4 repression in the ISR is suggested to be important among different physiological stresses.
C/EBPβ contributes to cell proliferation and differentiation and cellular stress responses (30–32). During the course of these processes, the activity of C/EBPβ can be regulated at multiple levels, including transcription, mRNA stability, protein phosphorylation, and translational control, that can lead to differential selection of start codons (30–32, 39, 40, 43, 49, 51–58). Consistent with these ideas, C/EBPβ mRNA is stabilized by UV irradiation (Fig. 5), and the levels of LIP protein can be transiently increased after UV irradiation (Figs. 6A and 10A) (43, 49, 51). C/EBPβ mRNA was reported to be stabilized by HuR, a protein that binds to AU-rich elements in the 3′-untranslated regions of mRNAs, which provides a mechanism for decreased decay of C/EBPβ transcripts in response to selected environmental stresses, such as UV irradiation (54). During the early phases of ER stress, up to about 3 h of thapsigargin, there is no comparable increase in LIP levels (Figs. 6 and 10A). This is consistent with prior studies, which reported no increase or in some cases a transient decrease in LIP protein levels during the first few hours of thapsigargin treatment of cultured cells (43, 49). Changes in C/EBPβ mRNA levels as well as translational control and stress signaling are likely to be central to the regulation of LIP repression of its target genes. Additionally, the availability of certain protein binding partners for the LIP bZIP transcription factor may be a contributing factor to the LIP repression of gene transcription.
Multiple Stress Response Pathways Can Contribute to ISR by Regulating ATF4 Transcription
C/EBPβ activity was reported to be regulated by different mitogen-activated protein kinases, mammalian target of rapamycin (mTOR); and eIF2α∼P (30, 39, 51, 59, 60). Therefore, a central target for cross-pathway regulation between the ISR and other stress response pathways is modulation of ATF4 transcription. In this way, eIF2α∼P and the ISR are not restricted to commensurate ATF4 synthesis, but rather the levels of ATF4 mRNA subject to preferential translation can be adjusted to the requirements of the cell for a specific environmental stress. This finding suggests that eIF2α∼P induced by a range of environmental stresses can lead to preferential translation of a differential subset of many different target genes. In the case of UV stress, there is repressed ATF4 expression, and induced eIF2α∼P can instead trigger preferential translation of alternative target mRNAs that facilitate DNA repair and enhance survival (Fig. 10A) (61). Selection of the precise target genes that are subject to preferential translation can be tailored to the individual stress condition, eliciting gene expression that is optimal for remedying the underlying cell damage.
Regarding transcriptional activation of ATF4, stresses that can increase ATF4 mRNA levels include ER stress (6, 29, 62), such as that induced by thapsigargin, starvation for amino acids (28), oxidative stress (63, 64), and certain anticancer agents (65, 66) (Fig. 10B). We are just beginning to understand the underlying mechanisms by which these stress conditions can induce ATF4 mRNA. In the case of oxidative stress, the transcription factor NRF2 was reported to bind to the ATF4 promoter and enhance its expression, which can serve to alleviate stress damage and facilitate angiogenesis (63, 64). The transcription factor CLOCK is suggested bind to the ATF4 promoter, resulting in enhanced ATF4 expression that can provide resistance to the anticancer drugs cisplatin and etoposide (67) (Fig. 10B). Finally, PDX1, a pancreas-specific transcription factor, activates the ATF4 promoter upon ER stress in islet β-cells (68). Together these findings suggest that many different transcription factors can bind to the ATF4 promoter and modulate the levels of ATF4 mRNA. Some of these transcription factors are inhibitors, triggering discordant induction of eIF2α∼P and ATF4 expression upon selected environmental stresses, whereas others are activators, amplifying ATF4 expression in the ISR. Furthermore, there can be tissue-specific regulation of ATF4 during certain stress conditions. As a consequence, multiple stress pathways can control the induction of ATF4 by eIF2α∼P, insuring that the levels of ATF4 and its ISR target genes are tailored for a given stress condition.
Acknowledgments
We thank Donghui Zhou for experimental assistance and acknowledge the contribution of Laura Zidek, who characterized the C/EBPβ-ΔuORF mice and generated MEF cells. Finally, we thank the Biochemistry Biotechnology Facility at Indiana University School of Medicine for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants GM049164 (to R. C. W.), DK053307 (to M. H.), and T32DK064466 (a predoctoral fellowship to B. F. T.). This work was also supported by American Heart Association Predoctoral Fellowship 11PRE7240012 (to B. F. T).
- eIF2∼P
- phosphorylation of eIF2
- C/EBPβ
- CCAAT/enhancer-binding protein β
- ISR
- integrated stress response
- LAP
- liver-enriched activating protein
- LIP
- liver-enriched inhibitory protein
- uORF
- upstream ORF
- bZIP
- basic leucine zipper
- ER
- endoplasmic reticulum
- MEF
- mouse embryonic fibroblast
- qPCR
- quantitative real time PCR
- TK
- thymidine kinase
- ASNS
- asparagine synthetase
- CAT-1
- cationic amino acid transporter 1
- Luc
- luciferase.
REFERENCES
- 1. Sonenberg N., Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 4. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 5. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 6. Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., Harding H. P. (2004) Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 9. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 10. Schröder M., Kaufman R. J. (2005) The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 11. Hinnebusch A. G., Natarajan K. (2002) Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 13. Jiang H. Y., Wek R. C. (2005) GCN2 phosphorylation of eIF2α activates NF-κB in response to UV irradiation. Biochem. J. 385, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng J., Harding H. P., Raught B., Gingras A. C., Berlanga J. J., Scheuner D., Kaufman R. J., Ron D., Sonenberg N. (2002) Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 12, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 15. Wek S. A., Zhu S., Wek R. C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15, 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gale M., Jr., Blakely C. M., Kwieciszewski B., Tan S. L., Dossett M., Tang N. M., Korth M. J., Polyak S. J., Gretch D. R., Katze M. G. (1998) Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18, 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sadler A. J. (2007) Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316, 253–292 [DOI] [PubMed] [Google Scholar]
- 18. García M. A., Meurs E. F., Esteban M. (2007) The dsRNA protein kinase PKR: virus and cell control. Biochimie 89, 799–811 [DOI] [PubMed] [Google Scholar]
- 19. Han A. P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S. H., Chen J. J. (2001) Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20, 6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J. J. (2007) Regulation of protein synthesis by the heme-regulated eIF2α kinase: relevance to anemias. Blood 109, 2693–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delépine M., Nicolino M., Barrett T., Golamaully M., Lathrop G. M., Julier C. (2000) EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 25, 406–409 [DOI] [PubMed] [Google Scholar]
- 22. Marciniak S. J., Ron D. (2006) Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 23. Pavitt G. D., Proud C. G. (2009) Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem. Soc. Trans. 37, 1298–1310 [DOI] [PubMed] [Google Scholar]
- 24. Bugiani M., Boor I., Powers J. M., Scheper G. C., van der Knaap M. S. (2010) Leukoencephalopathy with vanishing white matter: a review. J. Neuropathol. Exp. Neurol. 69, 987–996 [DOI] [PubMed] [Google Scholar]
- 25. Dey S., Baird T. D., Zhou D., Palam L. R., Spandau D. F., Wek R. C. (2010) Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33165–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar R., Krause G. S., Yoshida H., Mori K., DeGracia D. J. (2003) Dysfunction of the unfolded protein response during global brain ischemia and reperfusion. J. Cereb. Blood Flow Metab. 23, 462–471 [DOI] [PubMed] [Google Scholar]
- 27. Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. (2008) Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology 134, 568–576 [DOI] [PubMed] [Google Scholar]
- 28. Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. (2002) ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277, 24120–24127 [DOI] [PubMed] [Google Scholar]
- 29. Armstrong J. L., Flockhart R., Veal G. J., Lovat P. E., Redfern C. P. (2010) Regulation of endoplasmic reticulum stress-induced cell death by ATF4 in neuroectodermal tumor cells. J. Biol. Chem. 285, 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nerlov C. (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17, 318–324 [DOI] [PubMed] [Google Scholar]
- 31. Ramji D. P., Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsukada J., Yoshida Y., Kominato Y., Auron P. E. (2011) The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 54, 6–19 [DOI] [PubMed] [Google Scholar]
- 33. Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. (2005) Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 25, 59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 35. Siu F., Chen C., Zhong C., Kilberg M. S. (2001) CCAAT/enhancer-binding protein-β is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 276, 48100–48107 [DOI] [PubMed] [Google Scholar]
- 36. Bruhat A., Averous J., Carraro V., Zhong C., Reimold A. M., Kilberg M. S., Fafournoux P. (2002) Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 277, 48107–48114 [DOI] [PubMed] [Google Scholar]
- 37. Ma Y., Brewer J. W., Diehl J. A., Hendershot L. M. (2002) Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 38. Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. (1999) Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 39. Calkhoven C. F., Müller C., Leutz A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14, 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 40. Wethmar K., Bégay V., Smink J. J., Zaragoza K., Wiesenthal V., Dörken B., Calkhoven C. F., Leutz A. (2010) C/EBPβΔuORF mice—a genetic model for uORF-mediated translational control in mammals. Genes Dev. 24, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y., Bevilacqua E., Chiribau C. B., Majumder M., Wang C., Croniger C. M., Snider M. D., Johnson P. F., Hatzoglou M. (2008) Differential control of the CCAAT/enhancer-binding protein β (C/EBPβ) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J. Biol. Chem. 283, 22443–22456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., Wek R. C. (2008) Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 45. Descombes P., Schibler U. (1991) A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 46. Hatzoglou M., Fernandez J., Yaman I., Closs E. (2004) Regulation of cationic amino acid transport: the story of the CAT-1 transporter. Annu. Rev. Nutr. 24, 377–399 [DOI] [PubMed] [Google Scholar]
- 47. Schmitt-Ney M., Habener J. F. (2000) CHOP/GADD153 gene expression response to cellular stresses inhibited by prior exposure to ultraviolet light wavelength band C (UVC). Inhibitory sequence mediating the UVC response localized to exon 1. J. Biol. Chem. 275, 40839–40845 [DOI] [PubMed] [Google Scholar]
- 48. Wang X. Z., Lawson B., Brewer J. W., Zinszner H., Sanjay A., Mi L. J., Boorstein R., Kreibich G., Hendershot L. M., Ron D. (1996) Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 16, 4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chiribau C. B., Gaccioli F., Huang C. C., Yuan C. L., Hatzoglou M. (2010) Molecular symbiosis of CHOP and C/EBPβ isoform LIP contributes to endoplasmic reticulum stress-induced apoptosis. Mol. Cell. Biol. 30, 3722–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li B., Si J., DeWille J. W. (2008) Ultraviolet radiation (UVR) activates p38 MAP kinase and induces post-transcriptional stabilization of the C/EBPδ mRNA in G0 growth arrested mammary epithelial cells. J. Cell. Biochem. 103, 1657–1669 [DOI] [PubMed] [Google Scholar]
- 52. Chen C., Dudenhausen E. E., Pan Y. X., Zhong C., Kilberg M. S. (2004) Human CCAAT/enhancer-binding protein β gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J. Biol. Chem. 279, 27948–27956 [DOI] [PubMed] [Google Scholar]
- 53. Chen C., Dudenhausen E., Chen H., Pan Y. X., Gjymishka A., Kilberg M. S. (2005) Amino-acid limitation induces transcription from the human C/EBPβ gene via an enhancer activity located downstream of the protein coding sequence. Biochem. J. 391, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bergalet J., Fawal M., Lopez C., Desjobert C., Lamant L., Delsol G., Morello D., Espinos E. (2011) HuR-mediated control of C/EBPβ mRNA stability and translation in ALK-positive anaplastic large cell lymphomas. Mol. Cancer Res. 9, 485–496 [DOI] [PubMed] [Google Scholar]
- 55. Piwien-Pilipuk G., MacDougald O., Schwartz J. (2002) Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 277, 44557–44565 [DOI] [PubMed] [Google Scholar]
- 56. Timchenko L. T., Salisbury E., Wang G. L., Nguyen H., Albrecht J. H., Hershey J. W., Timchenko N. A. (2006) Age-specific CUGBP1-eIF2 complex increases translation of CCAAT/enhancer-binding protein β in old liver. J. Biol. Chem. 281, 32806–32819 [DOI] [PubMed] [Google Scholar]
- 57. Timchenko N. A., Wang G. L., Timchenko L. T. (2005) RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein β by interacting with the α and β subunits of eukaryotic initiation translation factor 2. J. Biol. Chem. 280, 20549–20557 [DOI] [PubMed] [Google Scholar]
- 58. Timchenko L. T., Iakova P., Welm A. L., Cai Z. J., Timchenko N. A. (2002) Calreticulin interacts with C/EBPα and C/EBPβ mRNAs and represses translation of C/EBP proteins. Mol. Cell. Biol. 22, 7242–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanlon M., Sturgill T. W., Sealy L. (2001) ERK2- and p90(Rsk2)-dependent pathways regulate the CCAAT/enhancer-binding protein-β interaction with serum response factor. J. Biol. Chem. 276, 38449–38456 [DOI] [PubMed] [Google Scholar]
- 60. Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. (2008) Dephosphorylation of translation initiation factor 2α enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7, 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Powley I. R., Kondrashov A., Young L. A., Dobbyn H. C., Hill K., Cannell I. G., Stoneley M., Kong Y. W., Cotes J. A., Smith G. C., Wek R., Hayes C., Gant T. W., Spriggs K. A., Bushell M., Willis A. E. (2009) Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 23, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 63. Afonyushkin T., Oskolkova O. V., Philippova M., Resink T. J., Erne P., Binder B. R., Bochkov V. N. (2010) Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler. Thromb. Vasc. Biol. 30, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 64. Miyamoto N., Izumi H., Miyamoto R., Bin H., Kondo H., Tawara A., Sasaguri Y., Kohno K. (2011) Transcriptional regulation of activating transcription factor 4 under oxidative stress in retinal pigment epithelial ARPE-19/HPV-16 cells. Invest. Ophthalmol. Vis. Sci. 52, 1226–1234 [DOI] [PubMed] [Google Scholar]
- 65. Levenson V. V., Davidovich I. A., Roninson I. B. (2000) Pleiotropic resistance to DNA-interactive drugs is associated with increased expression of genes involved in DNA replication, repair, and stress response. Cancer Res. 60, 5027–5030 [PubMed] [Google Scholar]
- 66. Tanabe M., Izumi H., Ise T., Higuchi S., Yamori T., Yasumoto K., Kohno K. (2003) Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res. 63, 8592–8595 [PubMed] [Google Scholar]
- 67. Igarashi T., Izumi H., Uchiumi T., Nishio K., Arao T., Tanabe M., Uramoto H., Sugio K., Yasumoto K., Sasaguri Y., Wang K. Y., Otsuji Y., Kohno K. (2007) Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene 26, 4749–4760 [DOI] [PubMed] [Google Scholar]
- 68. Sachdeva M. M., Claiborn K. C., Khoo C., Yang J., Groff D. N., Mirmira R. G., Stoffers D. A. (2009) Pdx1 (MODY4) regulates pancreatic β cell susceptibility to ER stress. Proc. Natl. Acad. Sci. U.S.A. 106, 19090–19095 [DOI] [PMC free article] [PubMed] [Google Scholar]