Fig. 1.
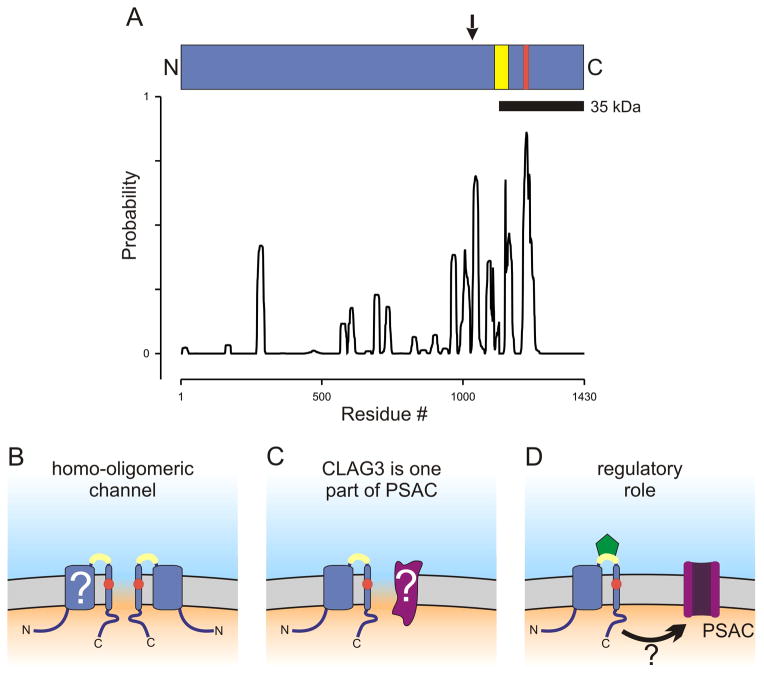
CLAG3 primary structure and possible contributions to PSAC formation. (A) Ribbon schematic showing the CLAG3 polypeptide aligned with a posterior probability plot for prediction of transmembrane domains, as calculated using Polyphobius. Arrow above the ribbon indicates site of recombination in the allelic exchange parasite, HB33rec. Yellow and red shading indicate variable residues and the site of a point mutation in a PSAC mutant, respectively. (B–D) Cartoons showing how erythrocyte membrane-embedded CLAG3 may contribute to PSAC formation and solute transport. The channel may be formed exclusively by CLAG3 oligomerization (panel B). The variable domain near the C-terminus is shown in yellow and is extracellular; a change in a PSAC mutant is indicated by a red circle within a predicted transmembrane segment. The transmembrane topology is not fully established (“?”). Panel C shows a heteromeric channel, where CLAG3 and an unidentified protein (purple, marked with “?”) both contribute to the channel pore. Panel D shows CLAG3 modulation of PSAC activity at a distance. ISPA-28 (green) is shown bound to the CLAG3 variable domain; how inhibitor binding is transmitted to the functional channel is unclear in this scenario (arrow with “?”).