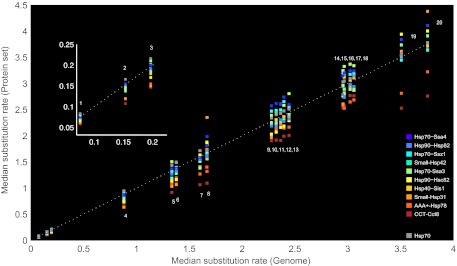
FIG. 3.—
Evolutionary distances of yeast substrates in the ten modules compared with their positional ortholog in 20 fungal species. The x axis shows the variation of amino acid substitution rates within different fungal genomes in comparison with yeast. The y axis shows the rate variation among proteins in the different modules within the same genome. Module colors correspond to the ranking by substrate expression levels with highly expressed modules in red shades and lowly expressed modules in blue shades. Hsp70 group includes five ungrouped chaperones: Ssb1, Ssa1, Sse1, Ssa2, and Ssb2. Arabic numerals correspond to fungal species: 1) Saccharomyces paradoxus, 2) Saccharomyces mikatae, 3) Saccharomyces bayanus, 4) Saccharomyces castellii, 5) Kluyveromyces Lactis, 6) Ashbya gossypii, 7) Kluyveromyces waltii, 8) Lachancea kluyveri, 9) Candida glabrata, 10) Candida guilliermondii, 11) Candida albicans, 12) Candida tropicalis, 13) Lodderomyces elongosporus, 14) Yarrowia lipolytica, 15) Aspergillus nidulans, 16) Neurospora crassa, 17) Schizosaccharomyces pompe, 18) Schizosaccharomyces japonicus, 19) Debaryomyces hansenii, and 20) Candida parapsilosis.