Summary
Restless Legs Syndrome (RLS), first chronicled by Willis in 1672 and described in more detail by Ekbom in 1945 [1], is a prevalent sensorimotor neurological disorder (5–10% in the population) with a circadian predilection for the evening and night. Characteristic clinical features also include a compelling urge to move during periods of rest, relief with movement, involuntary movements in sleep (viz., periodic leg movements of sleep), and fragmented sleep [2,3]. While the pathophysiology of RLS is unknown, dopaminergic neurotransmission and deficits in iron availability modulate expressivity [1,4–9]. GWAS have identified a polymorphism in an intronic region of the BTBD9 gene on chromosome 6 that confers substantial risk for RLS [2,3,10–12]. Here, we report that loss of the Drosophila homolog CG1826 (dBTBD9) appreciably disrupts sleep with concomitant increases in waking and motor activity. We further show that BTBD9 regulates brain dopamine levels in flies and controls iron homeostasis through the iron regulatory protein-2 (IRP2) in human cell lines. To our knowledge, this represents the first reverse genetic analyses of a “novel” or heretofore poorly understood gene implicated in an exceedingly common and complex sleep disorder and the development of an RLS animal model that closely recapitulates all disease phenotypes.
Results and Discussion
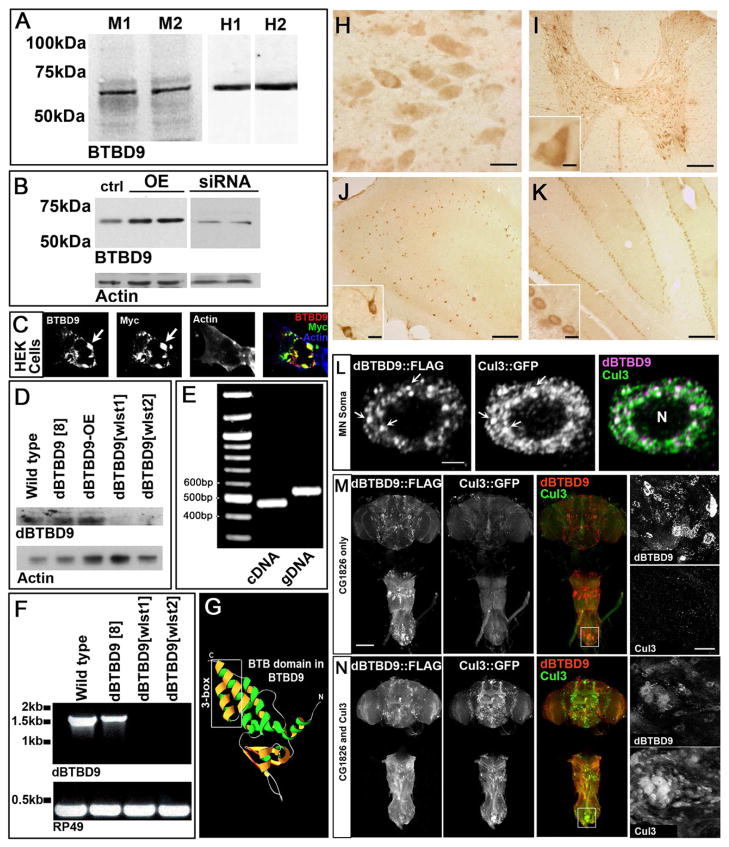
A number of genetic loci confer risk for RLS, including a SNP marker (rs3923809) in the gene BTBD9 that accounts for approximately 50% of the population attributable risk [11–16]. To confirm these findings and test the hypothesis that BTBD9 regulates sleep fragmentation, we examined the behavioral and metabolic consequences of disrupting BTBD9 function both in Drosophila and in human cell lines. BTBD9 is expressed widely in the mammalian nervous system and in HEK cell lines (Figures 1A and B) and is a predominantly a cytosolic protein (Figures 1C and H-K). A survey throughout the rat central nervous system reveals a widespread, yet specific, staining pattern for BTBD9 in circuits that regulate motor activity, memory, and emotion (Figures 1I, J and K; Figure S1A). Central nervous system expression is conserved in the Drosophila homolog dBTBD9 (CG1826) as seen with RT-PCR analysis of dBTBD9 mRNA and whole head western blots (Figures 1D and E). In the absence of reliable antibodies for dBTBD9 immunohistochemistry, we followed transgenically expressed FLAG-tagged full-length dBTBD9 and visualized Drosophila BTBD9 as distinct puncta in neuronal cytoplasm (Figure 1L and Figure S1B). Taken together, these results suggest that BTBD9 is a cytosolic protein that is widely expressed in the nervous system.
Figure 1. BTBD9 is a cytosolic Cullin-3 adaptor that is expressed widely in the mammalian and Drosophila nervous system.
(A) Western blots of two independent mouse and human brain protein extracts probed for BTBD9. (B) Protein extracts from HEK cells showing basal, over-expression and siRNA mediated knockdown of BTBD9. (C) HEK cell transfected with Myc-tagged BTBD9 and stained for BTBD9, Myc and Actin. Arrows indicate BTBD9 and Myc co-localization. (D) Western blots from Drosophila adult brain protein extracts probed for dBTBD9. dBTBD9[8] is a precise excision control, dBTBD9[OE] is pan-neuronal over-expression of dBTBD9 and dBTBD9[wlst1] and dBTBD9[wlst2] are null alleles. (E) PCR using cDNA and gDNA extracted from the adult Drosophila brain as template with intro-spanning primers for dBTBD9. (F) RT-PCR from mRNA isolated from control and excision alleles of dBTBD9. The ribosomal gene rp49 is used as a control. (G) Homology model of the N-terminal half of BTBD9 based on the crystal structure of SPOP showing the Cul-3 interacting 3-box. (H) BTBD9 staining in human nucleus basalis demonstrates cytoplasmic localization. Scale bar = 20μm. BTBD9 staining (I) throughout the grey matter of the rat spinal cord including motor neurons (inset shows enlarged view), (J) rat hippocampus (enlarged inset shows neurons of the CA1 region), and (K) Purkinje cells of the cerebellum (inset shows enlarged view). Scale bar = 200μm in (I–K) and 20μm (I–K insets). (L) A single neuron in the adult fly CNS stained for dBTBD9 (FLAG tagged dBTBD9) and Cul-3 (GFP tagged Cul-3) showing extensive co-localization (arrows). Scale bar = 2μm (M) CNS of an adult fly expressing FLAG tagged dBTBD9 (enlarged inset shows dBTBD9 aggregates). (N) CNS of a fly expressing both dBTBD9 and Cul-3, (inset shows enlarged view). Scale bar = 100μm. (See also Figure S1)
We hypothesized that BTBD9 belongs to a family of BTB domain containing substrate adaptors for the Cullin-3 (Cul-3) class of E3 Ubiquitin ligases [17] since homology modeling of both human and fly BTBD9 on the crystal structure of the MATH-BTB protein SPOP revealed the presence of a Cul-3 binding “3-box” motif [18] (Figure 1G). Consistent with this idea, FLAG tagged dBTBD9 shows extensive co-localization with Drosophila GFP tagged Cul-3 in neuronal soma (Figure 1L). Interestingly, over-expression of dBTBD9 without Cul-3 co-expression resulted in prominent aggregates that disappeared in the presence of excess Cul-3 (Figure 1M and N; Figures S3C and D). These data support BTBD9’s function as a Cul-3 adaptor and suggest that stringent stoichiometric ratios between BTBD9 and Cul-3 might be maintained under physiological conditions.
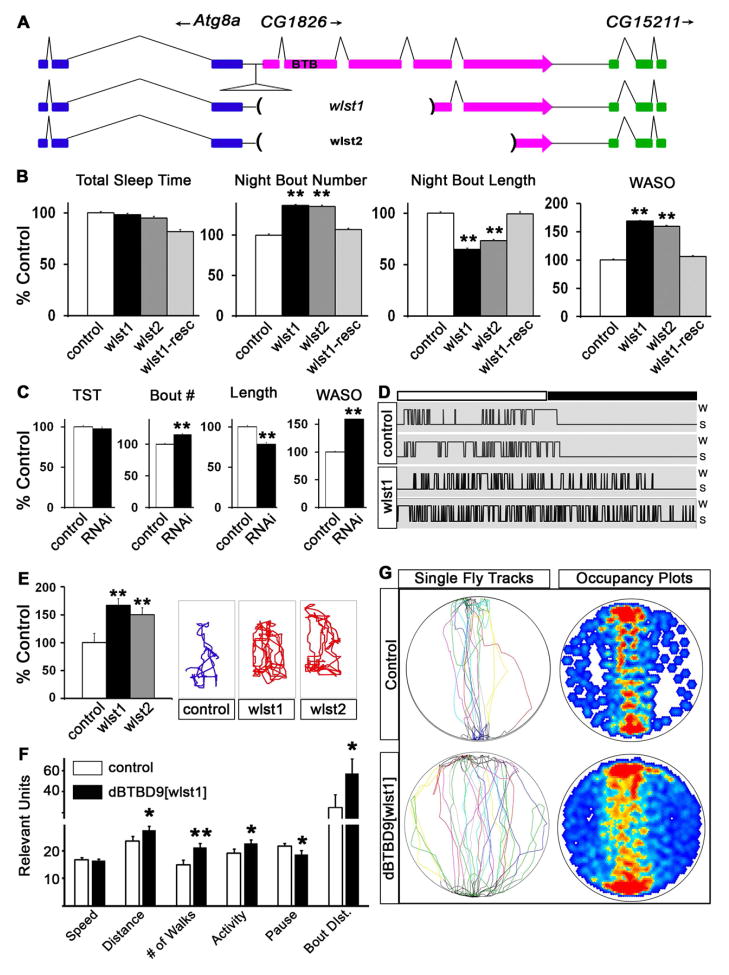
To examine phenotypic consequences resulting from loss of BTBD9, we isolated Drosophila mutants for dBTBD9 by excising a P-element transposon inserted 47 base pairs upstream of the dBTBD9 transcription start site (P{SUPor-P}CG1826KG07859) using standard genetic methods [19]. Two excision lines that carried large deletions in the dBTBD9 locus without affecting the adjacent genes Atg8a and CG15211, were selected for analysis (Figure 2A). These viable alleles, dBTBD9 wlst1 and dBTBD9 wlst2 (wlst is for “wanderlust” since these animals show increased locomotor activity, see below), did not produce any detectable dBTBD9 mRNA (Figure 1F) and were designated as null alleles. Both alleles had life spans that were significantly less than the genetically matched precise excision alleles that were used as controls suggesting fitness deficits that result from the loss of dBTBD9 (Figure S2B).
Figure 2. dBTBD9 mutants display sleep fragmentation and increased locomotor “restlessness”.
(A) Schematic of the dBTBD9 (CG1826) genomic region and also showing the extent of deletions in dBTBD9 null mutants. (B) Sleep phenotypes in two dBTBD9 null alleles (wlst1 and wlst2) compared with precise excision control and a pan-neuronal rescue with the wild type dBTBD9 transgene (wlst-resc). Total sleep time in a 24h period, mean number of sleep bouts at night, mean bout length at night and mean wake after sleep onset (WASO) are shown. More than 32 flies are tested for each genotype in this and other experiments. (C) Sleep phenotypes following pan-neuronal knockdown of dBTBD9 using a dBTBD9-specific RNAi and an elavC155-GAL4 driver. Total sleep time (TST), mean sleep bout number at night (Bout #), mean length of night-time sleep bout (Length) and WASO are shown. (D) Hypnograms, plots of sleep-wake transitions, over a 24 hour period are shown for two representative control and mutant (wlst1) animals. (E) The total distance walked by single flies within a restricted space (polycarbonate tube 5mm in external diameter, 10mm in length) in 1 minute. Representative tracks are shown for one animal for each genotype. (F, G) Control animals compared to dBTBD9[wlst1] animals in the Buridan’s assay. Speed of walking, total distance covered in 5 minutes, number of walks between the two black bars, total amount of time spent walking (activity), the number of pauses and the average length for a single uninterrupted walk (bout distance) are plotted. Single representative tracks as well as a heat map of the cumulative occupancy in the arena from 9 independent trials per genotype are shown. (p-values are: * <0.05; ** <0.01). All graphs are plotted as a percentage of control with error bars representing SEM. (See also Figure S2 and Movie S1)
Next, we monitored the Drosophila rest-activity cycle in dBTBD9 mutants using the Drosophila Activity Monitor (DAM) [20–22]. While the total duration of sleep per 24 hour period did not differ appreciably between control and mutant groups, the average bout lengths were decreased, the number of sleep bouts increased, and the amount of wake after sleep onset (WASO) increased (Figure 2B). Together, these phenotypes are emblematic of fragmented night-time sleep in dBTBD9 mutants, as demonstrated in hypnograms from individual flies (Figure 2D), which closely parallel the sleep continuity disruptions in RLS patients. These putative dBTBD9 mutations are allelic since they did not complement one another – rather, no sleep phenotypes were observed in heterozygotes (Figure S2A). Sleep phenotypes were unequivocally mapped to dBTBD9 since pan-neuronal add back of dBTBD9 (using the elavC155-GAL4 driver; [23,24]) in a null background rescued all sleep phenotypes (Figure 2B). Finally, RNA interference mediated knock down of dBTBD9 mRNA in the nervous system strongly phenocopied dBTBD9 null alleles (Figure 2C). These results clearly document sleep fragmentation in dBTBD9 mutants and indicate that neuronal dBTBD9 function is required for normal sleep architecture and consolidation in Drosophila.
A key endophenotype of RLS is the presence of Periodic Limb Movements (PLMs) in patients both during wakefulness and sleep [11,25–27]. To evaluate motor behavior in dBTBD9 mutants, we measured flight and negative geotaxis but these were normal (Figure S2C) [28]. However, when mutant flies were enclosed within a restricted space they were hyperlocomotive (Figure 2E and Supplementary Movie 1). This phenotype bears an uncanny resemblance to the “restlessness” observed in RLS patients asked to remain immobile in the “Suggested Immobilization Test” (SIT) [29]. Next, we turned to the visuomotor Buridan’s assay [30] to measure locomotion with greater analytical power. Such experiments revealed that mutant flies walked at the same speed as controls, but spent more time moving with fewer pauses (Figure 2F and G) resulting in longer uninterrupted bouts of walking. These experiments demonstrate that loss of dBTBD9, while not affecting general locomotion, upregulates the duration of motor activity, recapitulating the motor restlessness in RLS patients that ultimately impairs sleep consolidation.
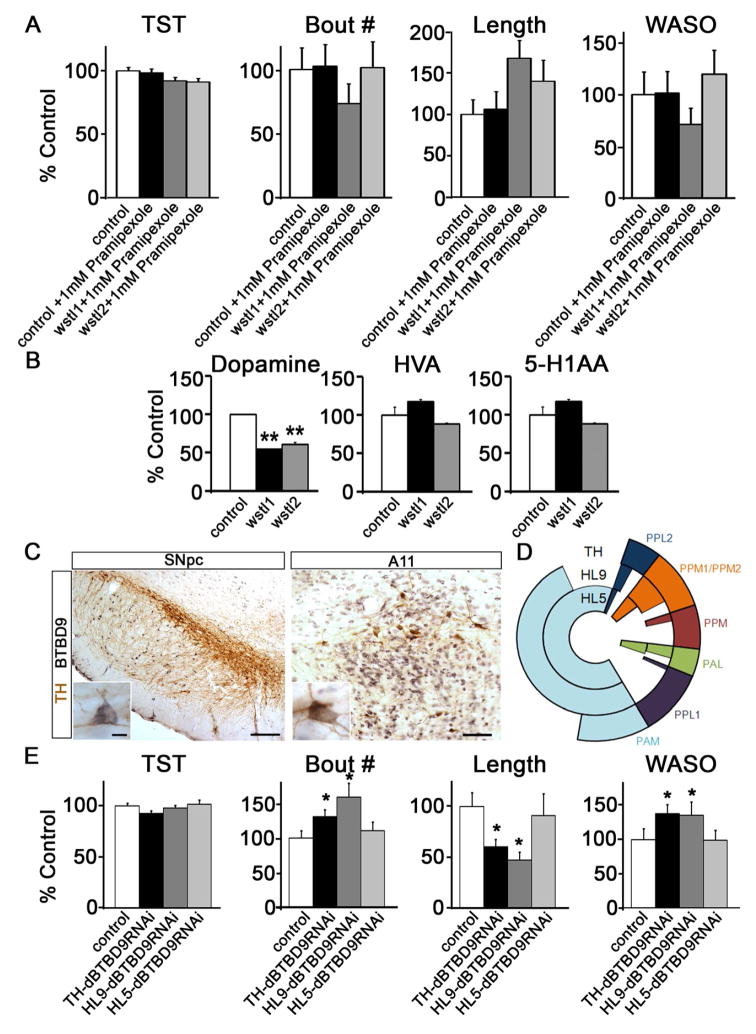
How does BTBD9 regulate sleep? Clinical information on RLS suggests aberrant dopamine signaling and iron homeostasis in RLS [3,31–34]. In the mammalian brain, BTBD9 is strongly expressed in dopaminergic neurons of the substantia nigra and A11 neurons (Figure 3C). Although we could not determine dBTBD9 expression in fly dopaminergic neurons, RNAi mediated knockdown of dBTBD9 in large subsets of dopaminergic neurons (using either the TH or HL9-GAL4 driver; [35, 36]) reproduced dBTBD9 mutant sleep fragmentation phenotypes (Figure 3D and E). This is consistent with expression of these two GAL4 drivers in dopaminergic neurons that are presumed to control locomotion [36]. Interestingly, we did not observe similar fragmentation when dBTBD9 was knocked down in a more restricted subset of dopaminergic neurons using the HL5-GAL4 line, a domain that does not influence ethanol-induced hyperactivity [37] (Figure 3D and E). Therefore, either BTBD9 mediates sleep phenotypes by altering neurotransmission in a small group of dopaminergic neurons targeted by both TH and HL9-GAL4, or dBTBD9 knockdown in a substantial number of dopaminergic neurons is required to induce sleep fragmentation. Given the central role of dopamine in sleep and arousal, we assessed dopamine levels in mutant and control fly brains using HPLC measurements. We found a 50% reduction in total dopamine that further affirmed a mechanistic link between dBTBD9 and dopamine (Figure 3B). A parsimonious explanation for reductions in dopamine includes reductions in tyrosine hydroxylase (TH), the rate limiting enzyme in dopamine biosynthesis, but this was not observed in dBTBD9 mutants (Figures S3A and B). However, these results do suggest that a principle mechanism by which BTBD9 modifies motor activity and sleep architecture is by ensuring normal dopamine biosynthesis. To test this idea, we augmented dopaminergic neurotransmission by feeding flies the non-ergoline dopamine agonist Pramipexole at a concentration of 1mM in normal fly food (Pramipexole preferentially targets dopamine D2-like receptors and is used clinically to treat RLS). Mutant flies treated with Pramipexole for 3 days showed marked improvement in sleep consolidation such that night time bout number, bout length and WASO were all rescued to control levels (Figure 3A). Note that the effect of Pramipexole increased incrementally with the duration of feeding (Figure S3C shows the difference in rescue between days 3 and 1 of Pramipexole feeding for controls and dBTBD9 mutants).
Figure 3. BTBD9 regulates dopaminergic neurotransmission to control sleep consolidation.
(A) Sleep phenotypes in dBTBD9 mutants when fed 1mM Pramipexole as compared to control animals. (B) HPLC based measurements of dopamine, homovanillic acid (HVA) and 5-hydroxyindoleacetic acid (5-HIAA) from control and dBTBD9 mutant adult fly heads. (p-values : ** <0.01). (C) Double labeled neurons expressing both tyrosine hydroxylase (DAB; brown) and BTBD9 (NiDAB; black) in the substantia nigra pars compacta (SNpc; Scale bar = 200μm) and A11 (Scale bar = 80μm). Enlarged images of double labeled neurons in each nucleus are shown in the insets. Scale bar = 20μm. (D) Schematic showing the type and number of dopaminergic neurons that are targeted by TH, HL9 and HL5-GAL4 lines. (E) Sleep phenotypes following RNAi mediated dBTBD9 knockdown in dopaminergic neurons using the TH-GAL4, HL9-GAL4 and HL5-GAL4 drivers. Total sleep time (TST), mean number of night-time sleep bouts (Bout #), mean night-time sleep bout length (Length) and WASO are shown. All graphs are plotted as a percentage of control with error bars representing SEM. (See also Figure S3).
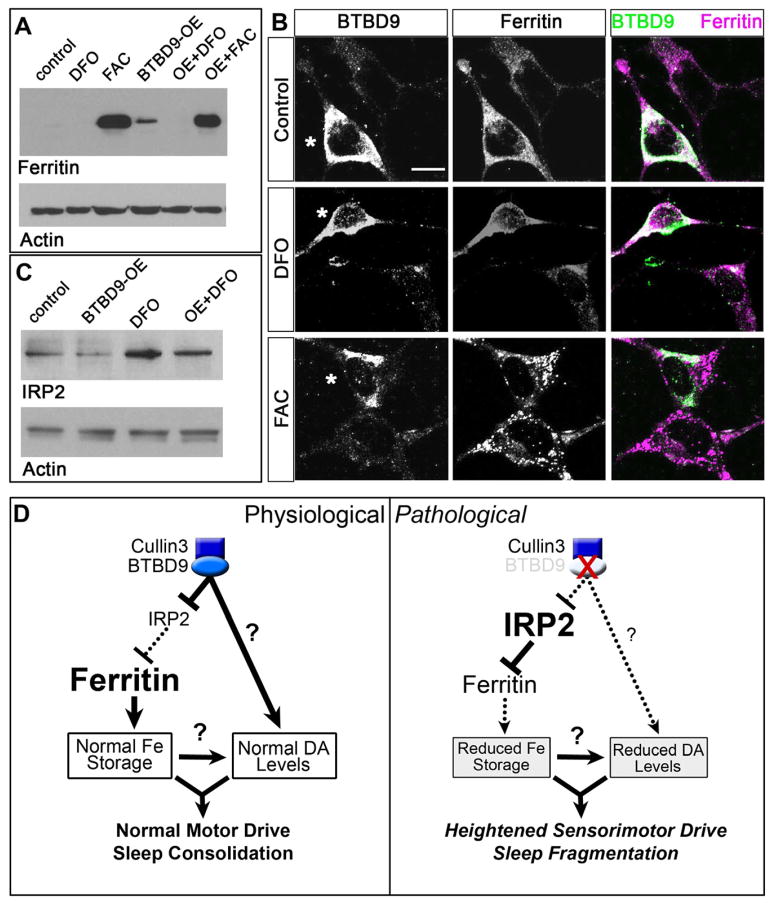
Low ferritin levels, reflecting impairments in the mobilization of iron stores, have been associated with RLS and the risk-conferring SNP in BTBD9 [10, 11]. Whether these findings implicate an extra-genetic role for iron, or rather, that BTBD9 influences iron metabolism is a critical question that has not been previously addressed [11,38]. Since investigation of iron metabolism in flies is in its infancy, we turned to HEK cells (which express BTBD9 endogenously) to determine the relationship between BTBD9 and iron storage. Over-expression of BTBD9 in HEK cells increased ferritin expression both under basal conditions and in the presence of the iron chelating siderophore deferoxamine mesylate (DFO) (Figures 4A and B). Mechanistically, the iron-responsive element-binding protein, IREB2 or IRP2 inhibits ferritin translation in response to changes in cellular iron, such that iron abundance leads to IRP2 degradation and a resultant increase in ferritin translation [39]. We therefore tested whether BTBD9 controls levels of IRP2. Western blots in Figure 4C show that both under basal and iron-chelation conditions, BTBD9 reduces IRP2 levels. This observation is consistent with the effect of BTBD9 on ferritin and profers a pathway by which BTBD9 modulates ferritin levels through regulation of IRP2 (Figure 4D). A relationship between BTBD9 and IRP2 also points to strongly conserved genetic and biochemical pathways that are central to a number of physiological contexts and pathological conditions and might also help to identify distinct causalities underlying RLS.
Figure 4. BTBD9 regulates iron metabolism by controlling ferritin expression via IRP2.
(A) Western blots of HEK cell protein lysates probed for ferritin light chain and actin (loading control). Ferritin levels are compared between untreated (control), deferoxamine mesylate (DFO) or ferric ammonium citrate (FAC) treated cells and BTBD9 transfected cells that are either untreated (BTBD9-OE) or treated with DFO or FAC. (B) BTBD9 and ferritin immunocytochemistry on HEK cells transfected with BTBD9 that are either untreated or treated with DFO or FAC. Asterisks mark BTBD9 transfected cells. Scale bar = 10 μm. (C) Western blot of HEK cell lysates probed for IRP2. DFO treated and untreated cells are compared with cells that are processed similarly but also over-express transgenic BTBD9. (D) Model of BTBD9 function under normal and pathological conditions. BTBD9 acts together with Cullin-3 to regulate IRP2 levels in the cell. This in turn controls ferritin expression and iron metabolism. Under normal conditions, BTBD9 inhibits IRP2 and promotes ferritin expression. BTBD9 also maintains dopamine biosynthesis either directly, or indirectly through iron, by currently unknown mechanisms. Together, this ensures normal motor drive and sleep consolidation. However, in RLS, loss of BTBD9 leads to increased IRP2, and reduced ferritin, thereby altering iron metabolism. It also negatively impacts dopamine synthesis leading to inappropriate motor activity and sleep fragmentation.
In sum, this study validates the GWAS for RLS by establishing powerful animal model of RLS in Drosophila based on the genetic risk factor BTBD9. Our results support the idea that genetic regulation of dopamine and iron metabolism constitute the core pathophysiology of at least some forms of RLS (Figure 4D). The extent to which this applies to RLS patients, and indeed the molecular nature of the human at-risk SNP in BTBD9, remain to be clarified. Together with the recent demonstration that Cullin-3 modulates sleep architecture in flies [40], our results further confirm and highlight the relevance of this pathway in sleep regulation. Finally, this study reiterates the utility of simple model systems such as Drosophila in unraveling the genetics of sleep and sleep disorders and opens up the possibility for querying additional phenotypes that are difficult to decipher in human populations.
Experimental Procedures
Drosophila husbandry, stocks, genetics and transgenesis
For stock information and genetics see supplementary experimental procedures. To create UAS-CG1826::FLAG animals, the CG1826 cDNA (DGRC gold clone) was cloned into the pENTR/D-TOPO entry vector (Invitrogen Inc.) followed by sequence verification and recombination into the pTWF destination vector. Transgenic animals were created by standard embryo microinjection (Bestgene Inc.).
Drosophila sleep measurements
Sleep measurements were carried out as described previously [20, 21] (for details see supplementary experimental procedures). Wake after sleep onset (WASO) was calculated as the amount of time the fly was active following the first sleep period after lights off (ZT12) until lights on (ZT0). Statistical significance was determined with an unpaired Student’s t-test and ANOVA.
Drosophila locomotor assays
Restricted space
Three to four day old flies were individually placed into the polycarbonate tubes used for sleep measurements (TriKinetics Inc.) and confined to a space 10mm in length with cotton plugs. After a 30 minute acclimation period, a series of five 1 minute videos were recorded with an HD webcam (Logitech.com). Movements were analyzed using the SpotTracker plugin for ImageJ and total distance travelled for each fly was averaged across the 5 trials.
Buridan’s assay
3 day old flies were collected and their wings cut close to the thorax. These flies were then allowed to recover for an additional 3 days. Flies were individually placed on a circular platform 10 cm in diameter and surrounded by a moat of water 2 cms in width. Flies normally walked back and forth between two diametrically opposite vertical black bars within a brightly illuminated cylinder. A camera mounted centrally above the platform recorded their movement for a period of 5 minutes. These recordings were then analyzed using custom designed software from Bjorn Brembs.
HPLC measurements and biochemistry
For HPLC measurements, fly heads were dissected from live animals under anesthesia, immediately frozen, and homogenized in chilled 0.1 M perchloric acid. After centrifugation, the supernatant was eluted through a C18 column (ESA Inc.) with the mobile phase containing 75 mM NaH2PO4, 1,5 mM Octanesulfonic acid and 5% Acetonitrile (pH 3.0). Western blotting was carried out following standard procedures (see supplementary experimental procedures). Primary antibody dilutions were as follows: mouse-anti-BTBD9 full-length (Abnova B01P, 1:1000), mouse-anti-actin (Millipore, 1:50,000), rabbit -anti-ferritin light chain (Abcam, 1:2000), mouse-anti-IRP2 (4G11) (Santa Cruz Biotechnology, 1:1000), rabbit-anti-TH (Wendi Neckameyer, 1:2000).
Anatomy, immunohistochemistry and microscopy
Mammalian and Drosophila neuroanatomy
Anatomical experiments were carried out on fixed tissue followed by either brightfield or laser scanning confocal microscopy (see supplementary experimental procedures for details). Antibody dilutions were as follows: mouse anti-BTBD9 1:1000 (Abnova), mouse anti-Elav 1:25 (DSHB), Phalloidin 543 (Molecular Probes 1:50), rabbit anti-GFP 1:400 (Invitrogen) and rabbit anti-TH (1:1000) (Wendi Neckameyer). Secondary antibody dilutions are as follows: Anti-Rabbit IgG-Alexa Flour 488, 563 (MolecularProbes 1:500), anti-mouse IgG-Alexa Flour 488, 568 (MolecularProbes 1:500).
Statistical analysis
Unpaired Student’s t-test and ANOVA was used for statistical analysis of sleep and locomotion data.
Supplementary Material
Highlights.
The RLS risk factor BTBD9 regulates sleep in Drosophila
BTBD9 mutants display sleep fragmentation and increased motor restlessness
BTBD9 regulates dopamine levels and iron homeostasis
We present the first reverse genetic model of a human sleep disorder in Drosophila
Acknowledgments
We thank members of the Betarbet, Jinnah, Rye and Sanyal laboratory for technical assistance, comments and criticisms, Diana Woodall and Sonya Patel for initial characterization of fly phenotypes, Paul Shaw for sharing his sleep analysis software, Bjorn Brembs for the Buridan analysis software and Wendi Neckameyer for sharing the anti-TH antibody. This work is supported by a FIRST fellowship and a Neurology NIH T32 fellowship to A.F. and Emory Neuroscience Initiative, Sleep Research Society and Restless Legs Foundation grants to S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ekbom KA. Restless legs. Acta Med Scand. 1945;158:1–123. [Google Scholar]
- 2.Trotti LM, Rye DB. Restless legs syndrome. Handb Clin Neurol. 2011;100:661–73. doi: 10.1016/B978-0-444-52014-2.00047-1. [DOI] [PubMed] [Google Scholar]
- 3.Trenkwalder C, Paulus W. Restless legs syndrome: pathophysiology, clinical presentation and management. Nat Rev Neurol. 2010;6:337–46. doi: 10.1038/nrneurol.2010.55. [DOI] [PubMed] [Google Scholar]
- 4.Allen RP, Earley CJ. The role of iron in restless legs syndrome. Mov Disord. 2007;22 (Suppl 18):S440–8. doi: 10.1002/mds.21607. [DOI] [PubMed] [Google Scholar]
- 5.Aul EA, Davis BJ, Rodnitzky RL. The importance of formal serum iron studies in the assessment of restless legs syndrome. Neurology. 1998;51:912. doi: 10.1212/wnl.51.3.912. [DOI] [PubMed] [Google Scholar]
- 6.Sun ER, Chen CA, Ho G, Earley CJ, Allen RP. Iron and the restless legs syndrome. Sleep. 1998;21:371–7. [PubMed] [Google Scholar]
- 7.Montplaisir J, Lorrain D, Godbout R. Restless legs syndrome and periodic leg movements in sleep: the primary role of dopaminergic mechanism. Eur Neurol. 1991;31:41–3. doi: 10.1159/000116643. [DOI] [PubMed] [Google Scholar]
- 8.Hening WA. Restless Legs Syndrome. Curr Treat Options Neurol. 1999;1:309–319. doi: 10.1007/s11940-999-0021-9. [DOI] [PubMed] [Google Scholar]
- 9.Turjanski N, Lees AJ, Brooks DJ. Striatal dopaminergic function in restless legs syndrome: 18F-dopa and 11C-raclopride PET studies. Neurology. 1999;52:932–7. doi: 10.1212/wnl.52.5.932. [DOI] [PubMed] [Google Scholar]
- 10.Allen R. Dopamine and iron in the pathophysiology of restless legs syndrome (RLS) Sleep Med. 2004;5:385–91. doi: 10.1016/j.sleep.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Stefansson H, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 12.Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 13.Kemlink D, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. 2009;46:315–8. doi: 10.1136/jmg.2008.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, et al. Association studies of variants in MEIS1, BTBD9, and MAP2K5/SKOR1 with restless legs syndrome in a US population. Sleep Med. 2011;12:800–4. doi: 10.1016/j.sleep.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schormair B, et al. MEIS1 and BTBD9: genetic association with restless leg syndrome in end stage renal disease. J Med Genet. 2011;48:462–6. doi: 10.1136/jmg.2010.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Diaz H, Iranzo A, Rye DB, Santamaria J. Restless abdomen: a phenotypic variant of restless legs syndrome. Neurology. 2011;77:1283–6. doi: 10.1212/WNL.0b013e318230207a. [DOI] [PubMed] [Google Scholar]
- 17.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul-3-BTB complexes join the family. EMBO J. 2004;23:1681–7. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul-3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–94. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 20.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 21.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 22.Agosto J, et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–9. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 24.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 25.Trenkwalder C, Walters AS, Hening W. Periodic limb movements and restless legs syndrome. Neurol Clin. 1996;14:629–50. doi: 10.1016/s0733-8619(05)70277-2. [DOI] [PubMed] [Google Scholar]
- 26.Vetrugno R, Provini F, Montagna P. Restless legs syndrome and periodic limb movements. Rev Neurol Dis. 2006;3:61–70. [PubMed] [Google Scholar]
- 27.Coleman RM, Pollak CP, Weitzman ED. Periodic movements in sleep (nocturnal myoclonus): relation to sleep disorders. Ann Neurol. 1980;8:416–21. doi: 10.1002/ana.410080413. [DOI] [PubMed] [Google Scholar]
- 28.Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc Natl Acad Sci U S A. 1967;58:1112–9. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montplaisir J, et al. Immobilization tests and periodic leg movements in sleep for the diagnosis of restless leg syndrome. Mov Disord. 1998;13:324–9. doi: 10.1002/mds.870130220. [DOI] [PubMed] [Google Scholar]
- 30.Gotz KG. Visual guidance in Drosophila. Basic Life Sci. 1980;16:391–407. doi: 10.1007/978-1-4684-7968-3_28. [DOI] [PubMed] [Google Scholar]
- 31.Clemens S, Rye D, Hochman S. Restless legs syndrome: revisiting the dopamine hypothesis from the spinal cord perspective. Neurology. 2006;67:125–30. doi: 10.1212/01.wnl.0000223316.53428.c9. [DOI] [PubMed] [Google Scholar]
- 32.Varga LI, et al. Critical review of ropinirole and pramipexole - putative dopamine D(3)-receptor selective agonists - for the treatment of RLS. J Clin Pharm Ther. 2009;34:493–505. doi: 10.1111/j.1365-2710.2009.01025.x. [DOI] [PubMed] [Google Scholar]
- 33.Earley CJ, et al. The dopamine transporter is decreased in the striatum of subjects with restless legs syndrome. Sleep. 2011;34:341–7. doi: 10.1093/sleep/34.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montplaisir J, Godbout R, Poirier G, Bedard MA. Restless legs syndrome and periodic movements in sleep: physiopathology and treatment with L-dopa. Clin Neuropharmacol. 1986;9:456–63. doi: 10.1097/00002826-198610000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Friggi-Grelin F, et al. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–27. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 36.Claridge-Chang A, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–15. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong EC, et al. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS One. 2010;5:e9954. doi: 10.1371/journal.pone.0009954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Earley CJ, et al. Abnormalities in CSF concentrations of ferritin and transferrin in restless legs syndrome. Neurology. 2000;54:1698–700. doi: 10.1212/wnl.54.8.1698. [DOI] [PubMed] [Google Scholar]
- 39.Samaniego F, Chin J, Iwai K, Rouault TA, Klausner RD. Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. Structure, function, and post-translational regulation. J Biol Chem. 1994;269:30904–10. [PubMed] [Google Scholar]
- 40.Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–76. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.