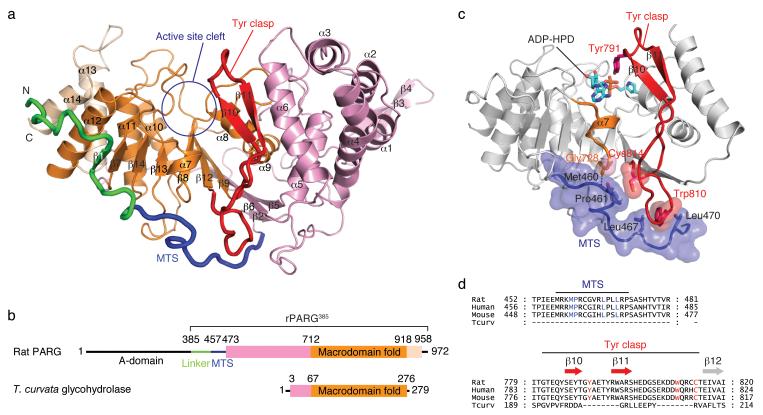
Figure 1.
Mammalian poly(ADP-ribose) glycohydrolase (PARG) structure. (a) The catalytic domain of rat PARG (rPARG385; residues 385-972) consists of a core macrodomain fold (orange) sandwiched between flanking N-terminal (pink) and C-terminal (beige) helical bundles. The mitochondrial targeting sequence (MTS; blue) wraps around the catalytic domain and stabilizes the tyrosine clasp (Tyr clasp; red), a unique substrate-binding element of mammalian PARG. (b) Domain organization of rat PARG and Thermomonospora curvata glycohydrolase. (c) The MTS buttresses the Tyr clasp, orienting Tyr791 towards the active site cleft, explaining why the MTS is required for PARG activity. (d) Sequence alignment reveals conserved residues in mammalian PARG that participate in the interaction between the MTS and the Tyr clasp.