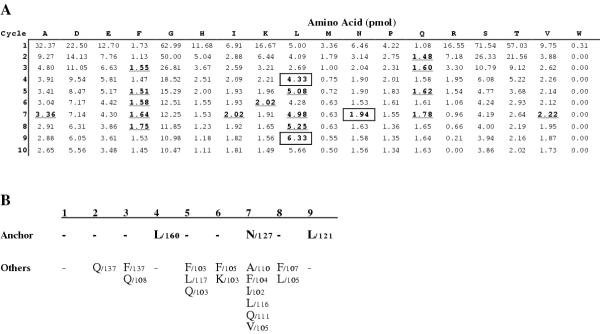
Figure 4.
Peptide binding motif obtained by refolding recombinant HLA-E around a nonamer random peptide library. Denatured recombinant HLA-E heavy chain was refolded around a 100-fold molar excess of nonamer random peptides in the presence of a 2-fold excess of β2-microglobulin. Complexes were purified by gel filtration and their bound peptides released by acid elution. This population was purified further by reversed-phase chromatography and sequenced by Edman degradation. A Results show the yield of each amino acid at each cycle. Anchor residues were determined as described in the "Materials and Methods" and are boxed whilst other increases are underlined. B The peptide motif of the pool sequence data from A. Values next to residues are the % increases compared to the previous sequencing cycle.