Figure 1.
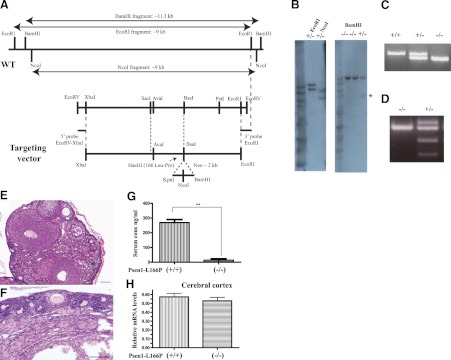
Generation and characterization of Psen1-L166P-knock-in mice. A) Schematic representation of the Psen1 WT allele and targeting vector. The codon encoding the L166P mutation is located in exon 6. A SacI-PstI (2.1 kb) subclone was mutagenized to change Leu to Pro, which created a HaeIII site. The AvaI-BsaI (540 bp) fragment containing the mutation was sequenced to ensure that no PCR error was introduced. This fragment was used in the final targeting vector construction. The targeting vector was linearized with NotI and purified before C57Bl/6J ES cells were electrophorated. Neomycin-resistant ES clones were selected. B) Southern blot analysis shows homologous recombination in both arms in DNA from selected ES cells digested with EcoRI and NcoI and hybridized with the 5′ EcoRV-XbaI (∼0.3 kb) probe. The WT allele is ∼9 kb, and the knock-in (KI) allele is ∼11 kb in EcoRI-digested DNA and ∼9 and ∼6 kb in NcoI-digested DNA, respectively. Tail genomic DNA was digested with BamHI and hybridized with the 3′EcoRI probe. The WT allele is ∼11 kb, and the KI allele is ∼5 kb (asterisk). C) Genotyping by PCR. The top band is amplified from the mutant allele (670 bp), and the bottom band from the wild-type allele (558 bp). A single band (670 bp) is seen in homozygous Psen1-L166P (+/+) mice, and a single band (558 bp) is seen in WT Psen1-L166P (−/−) mice. D) Restriction enzyme analysis of PCR-amplified genomic DNA using HaeIII. WT allele (−/−) cannot be digested. Digestion is only observed when the mutant allele (+/−) is present, confirming the presence of the L166P mutation. E, F) Hematoxylin and eosin-stained sections of ovaries of 9-mo-old female normal (−/−) (E) and homozygous Psen1-L166P (+/+) (F) mice. Numerous follicles at different stages can be seen throughout the normal ovary (E), but not in (+/+) mice (F). Scale bars = 100 μm. G) Serum concentrations of FSH in 3–4-mo-old female homozygous Psen1-L166P (+/+) mice and normal Psen1-L166P (−/−) mice. **P < 0.001. H) Multiplex RT-PCR expression analysis. Bar graphs depict differential gene expression levels between homozygous Psen1-L166P (+/+) mice and normal Psen1-L166P (−/−) mice. Multiplex RT-PCR analysis was performed on mRNA isolated from the cerebral cortex. Analysis was performed in triplicate and normalized to the Polr2a gene. Group averages are reported as relative mRNA levels (means±sd). No significant expression differences were found (P<0.05) by 2-tailed t test.