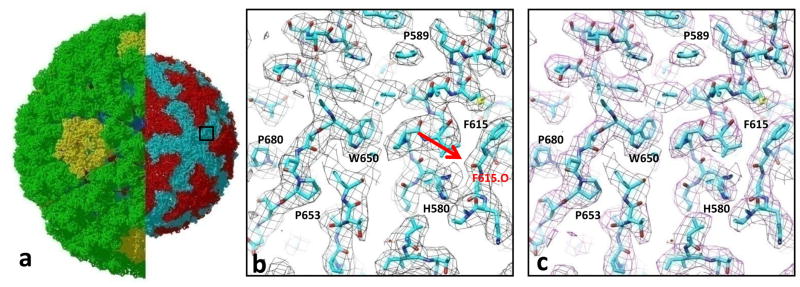
Figure 4. Accuracy of the cryoEM density map computed by eLite3D.
(a) Atomic model of the GCRV ISVP derived from a 3.3Å 3D reconstruction. The left half shows both its coat and core. In the right half, the coat shell is removed to reveal the core. The box demarcates the region displayed in (b) and (c). (b) and (c) Close-up views of the boxed area in (a), showing cryoEM densities (mess) calculated using the eLite3D GPGPU software (b) and serial CPU software(Frealign) (c). The atomic models are shown using sticks with numbers indicating amino-acid residues. Sidechain densities of the core protein are clearly resolved in both maps, and some of the carboxyl oxygen atoms are also resolved [e.g., the carboxyl oxygen of F615, arrow in (b)].