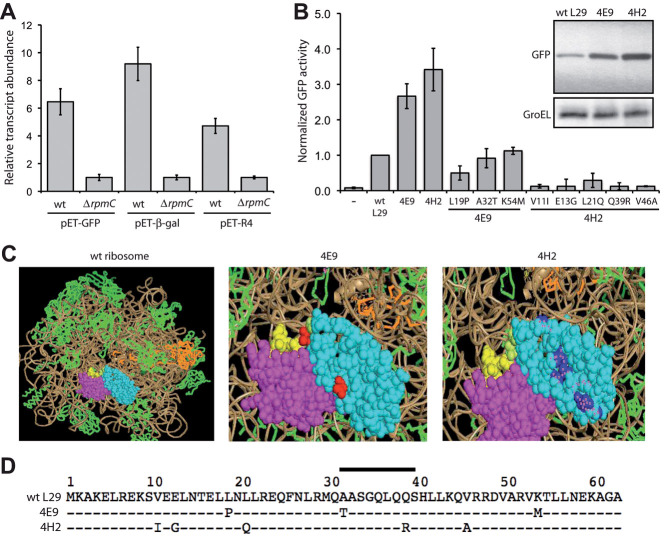
Figure 2.
Characterization and engineering of GFP expression in L29-deficient cells. (A) qRT-PCR analysis of mRNA transcript levels for GFP, β-gal and scFv13-R4. RNA was isolated from wt and ΔrpmC::kan cells carrying the plasmid indicated. All data was normalized to the amount of 16S rRNA measured in each strain. Relative induction was calculated by dividing each normalized value by the value measured in ΔrpmC::kan cells expressing GFP, β-gal, or scFv13-R4. Data show the average of six independent experimental repeats and the error bars represent the SEM. (B) Cellular GFP activity produced by ΔrpmC::kan cells in the presence of empty plasmid (–), wt L29, or the L29 variants 4E9, 4H2, and the single amino acid substitution mutants as indicated. Cell fluorescence was measured by FACS and cells were ungated. Values were normalized to control cells co-expressing GFP and wt L29. Data show the average of three independent experimental repeats and the error bars represent the SEM. Inset shows western blot analysis of whole cell lysates isolated from ΔrpmC::kan cells co-expressing GFP and different L29 variants as indicated. GroEL served as a loading control. (C) Structural depiction of wt and mutant L29 proteins (cyan) in the context of L23 (magenta) and the TF-binding site (yellow). Mutations in 4E9 and 4H2 are shown in red and blue, respectively. (D) Amino acid sequence alignment of E. coli L29 and related variants. Bar indicates the TF-binding region.