Figure 6.
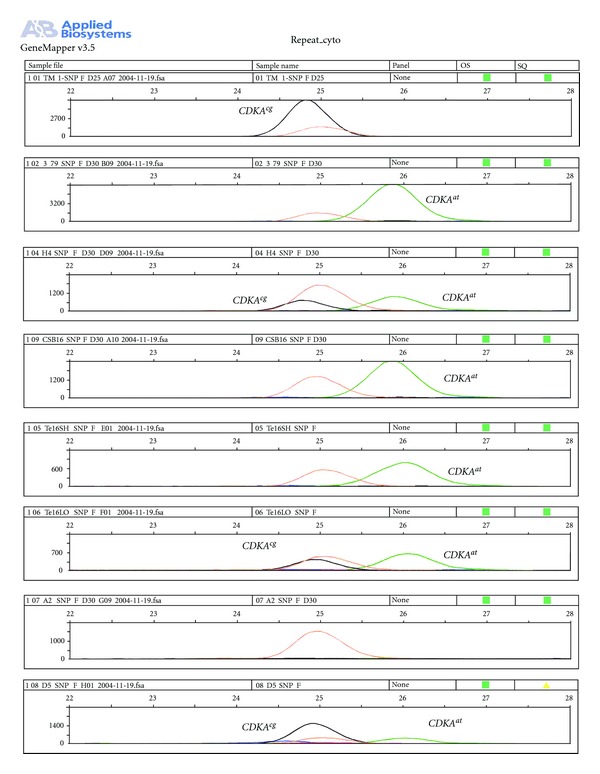
The electropherograms of two allelic SNPs, designated here as CDKAcg (black) and CDKAat (green), that corresponded to the polymorphism between G. hirsutum inbred TM-1 and G. barbadense-doubled haploid 3–79. Genomic dosage profiles are shown for [A] TM-1; [B] 3–79; [C–E] three hypoaneuploid-interspecific G. hirsutum x G. barbadense F1 hybrids, [C] lacking G. hirsutum chromosome 4 (H4), [D] a monotelodisomic-16sh F1 (Te16SH) lacking most of the long arm of G. hirsutum chromosome 16, [E] a monotelodisomic 16 Lo F1 (Te16LO) lacking most of the short arm of G. hirsutum chromosome-16; [F] a backcross disomic substitution plant (CSB-16) in which chromosome 16 of 3–79 has replaced the TM-1 chromosome 16; [G] a G. arboreum (A2 species); and [H] a G. raimondii plant (D5 species). Electropherograms revealing both peaks that indicate heterozygosity for both parental SNPs, as in the H4 interspecific F1 hybrid, indicate the locus is not in this chromosome. In contrast, absence of the G. hirsutum SNP CDKAcg from CSB 16 and Te16SH hybrids and its presence in Te16LO concordantly indicate that the SNP marker is located in the long arm of the chromosome 16.
