Figure 5.
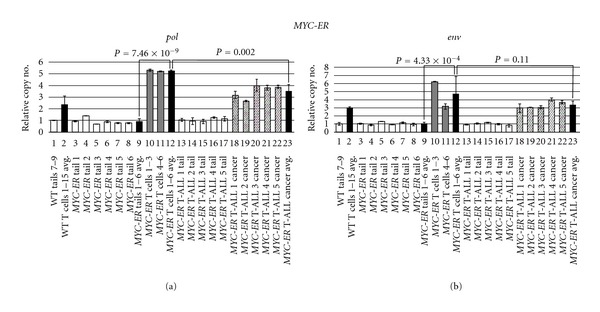
ZFERV amplifications in MYC-ER zebrafish. Phenotypically normal (n = 6) or T-ALL+ (n = 5) MYC-ER fish were tested by qPCR of pol (a) and env (b). White bars display tail DNA from single normal (lanes 3–8) or diseased (lanes 13–17) fish. MYC-ER tails had similar copy number to each other (compare lanes 3–8 and 13–17) and to WT Tails 7–9 (lane 1). T cells pooled from groups of 3 normal MYC-ER fish (gray bars) showed ZFERV amplification; higher gains were seen in MYC-ER than WT T cells (lane 2 versus 12; P values 5.86 × 10−4 for pol, 0.15 for env). T cells showed 4- to 5-fold higher ZFERV copy than matched tails (lane 9 versus 12). Diagonally hatched bars depict amplifications in T-ALL cells from 5 MYC-ER fish (lanes 18–22). Cancer ZFERV levels were well above paired tails (compare lanes 13–17 to 18–22). Slightly lower gains were seen in cancerous than non-malignant MYC-ER T cells (lane 12 versus 23); this reached statistical confidence for pol, but not env. Other details are as listed in Figure 3's legend.
