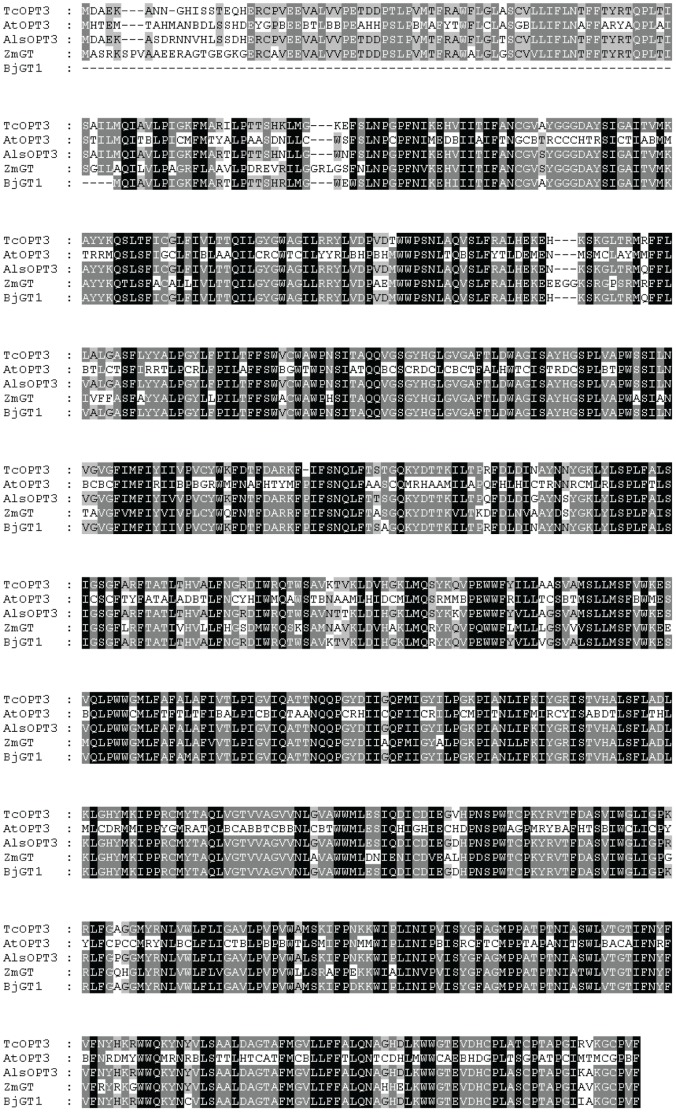
Figure 1. Sequence analysis of TcOPT3.
Sequence alignment among the TcOPT3, AtOPT3 ATOPT3, ZmGT and BjGT1. CLUSTAL W (version 1.8) alignment of deduced amino acid sequences from the OPTs. Amino acids are numbered from the initiator ATG. Black-shaded areas represent the consensus, dark-gray-shaded areas represent identical amino acids, and light-gray-shaded areas represent similar amino acids. The putative transmembrane (TM) domains of the TcOPT3 were determined by the TMHMM algorithm. The predicted transmembrane membrane spanning domains are shown as lines above the sequences, and numbered TM I–TM XIV respectively. The bars under the sequence show the location of the two conserved motifs (NPG and KIPPR motifs). Tc, T. caerulescens; At, Arabidopsis thaliana; Als, Arabidopsis lyrata subsp.; Zm, Zea mays; Bj, Brassica juncea. Accession numbers are: HQ699884 (TcOPT3), NP_567493 (AtOPT3), XP_002868139 (AlsOPT3), ACL82964 (ZmGT), CAD91127.1 (BjGT1).