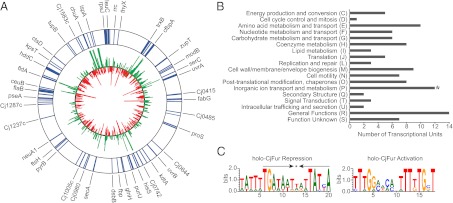
Fig. 1.
(A) Genomic map of CjFur-enriched transcriptional units overlaid with C. jejuni’s transcriptional response to iron limitation. The outer ring lists all of the transcriptional units that were enriched under Fur ChIP (≥1.5 enrichment, P ≤ 10−4). Blue denotes the gene that was found to be enriched under Fur ChIP. The inner ring displays the transcriptional response of each gene to iron limitation, with green denoting iron-repressed genes and red denoting iron-activated genes (≥1.5 fold change, P ≤ 10−4). The figure was made using Circos version 0.54 (34). (B) COG functional groups present in CjFur ChIP-enriched transcriptional units. CjFur ChIP-enriched genes encompass a diverse range of COG functional categories, indicating that CjFur plays regulatory roles beyond iron metabolism. The COG functional category inorganic ion transport and metabolism was found to be statistically overrepresented (*). (C) Consensus sequences for CjFur binding sites. (Left) Holo-CjFur repression consensus sequence, with its palindromic sequence highlighted with arrows. (Right) Holo-CjFur activation consensus sequence. Consensus sequences were made using WebLogo 3 (http://weblogo.berkeley.edu).